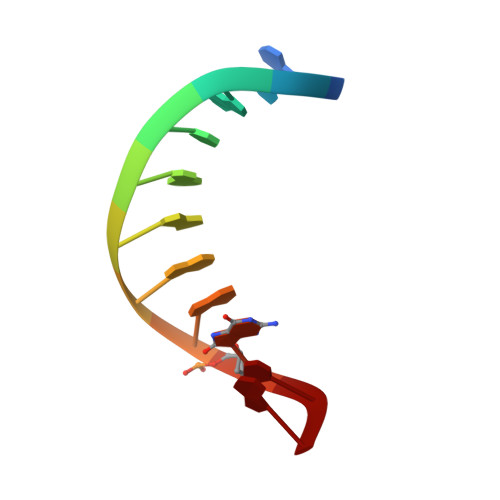
Structure and dynamics of DNA duplexes containing a cluster of mutagenic 8-oxoguanine and abasic site lesions.
Zalesak, J., Lourdin, M., Krejc, L., Constant, J.F., Jourdan, M.(2014) J Mol Biol 426: 1524-1538
- PubMed: 24384094
- DOI: https://doi.org/10.1016/j.jmb.2013.12.022
- Primary Citation of Related Structures:
2M3P, 2M3Y, 2M40, 2M43, 2M44 - PubMed Abstract:
Clustered DNA damage sites are caused by ionizing radiation. They are much more difficult to repair than are isolated single lesions, and their biological outcomes in terms of mutagenesis and repair inhibition are strongly dependent on the type, relative position and orientation of the lesions present in the cluster. To determine whether these effects on repair mechanism could be due to local structural properties within DNA, we used (1)H NMR spectroscopy and restrained molecular dynamics simulation to elucidate the structures of three DNA duplexes containing bistranded clusters of lesions. Each DNA sequence contained an abasic site in the middle of one strand and differed by the relative position of the 8-oxoguanine, staggered on either the 3' or the 5' side of the complementary strand. Their repair by base excision repair protein Fpg was either complete or inhibited. All the studied damaged DNA duplexes adopt an overall B-form conformation and the damaged residues remain intrahelical. No striking deformations of the DNA chain have been observed as a result of close proximity of the lesions. These results rule out the possibility that differential recognition of clustered DNA lesions by the Fpg protein could be due to changes in the DNA's structural features induced by those lesions and provide new insight into the Fpg recognition process.
Organizational Affiliation:
Département de Chimie Moléculaire, CNRS UMR5250, ICMG FR2607, Université de Grenoble, 570 rue de la Chimie, BP 53, 38041 Grenoble Cedex 9, France.