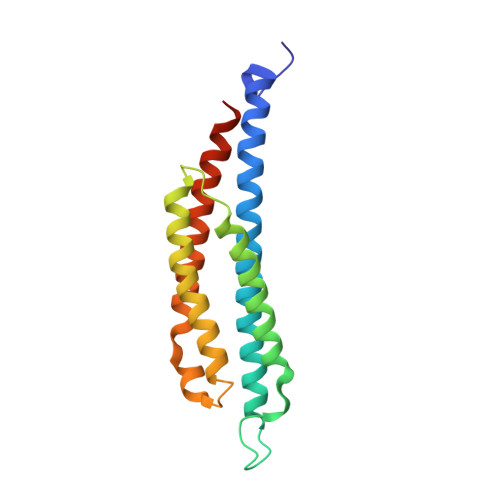
Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand.
Milburn, M.V., Prive, G.G., Milligan, D.L., Scott, W.G., Yeh, J., Jancarik, J., Koshland Jr., D.E., Kim, S.H.(1991) Science 254: 1342-1347
- PubMed: 1660187
- DOI: https://doi.org/10.1126/science.1660187
- Primary Citation of Related Structures:
1LIH, 2LIG - PubMed Abstract:
The three-dimensional structure of an active, disulfide cross-linked dimer of the ligand-binding domain of the Salmonella typhimurium aspartate receptor and that of an aspartate complex have been determined by x-ray crystallographic methods at 2.4 and 2.0 angstrom (A) resolution, respectively. A single subunit is a four-alpha-helix bundle with two long amino-terminal and carboxyl-terminal helices and two shorter helices that form a cylinder 20 A in diameter and more than 70 A long. The two subunits in the disulfide-bonded dimer are related by a crystallographic twofold axis in the apo structure, but by a noncrystallographic twofold axis in the aspartate complex structure. The latter structure reveals that the ligand binding site is located more than 60 A from the presumed membrane surface and is at the interface of the two subunits. Aspartate binds between two alpha helices from one subunit and one alpha helix from the other in a highly charged pocket formed by three arginines. The comparison of the apo and aspartate complex structures shows only small structural changes in the individual subunits, except for one loop region that is disordered, but the subunits appear to change orientation relative to each other. The structures of the two forms of this protein provide a step toward understanding the mechanisms of transmembrane signaling.
Organizational Affiliation:
Department of Chemistry, University of California, Berkeley 94720.