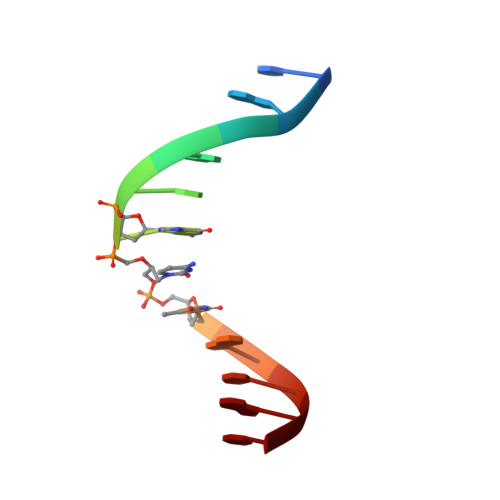
Solution structure of a DNA duplex containing the potent anti-poxvirus agent cidofovir.
Julien, O., Beadle, J.R., Magee, W.C., Chatterjee, S., Hostetler, K.Y., Evans, D.H., Sykes, B.D.(2011) J Am Chem Soc 133: 2264-2274
- PubMed: 21280608
- DOI: https://doi.org/10.1021/ja109823e
- Primary Citation of Related Structures:
2L8P, 2L8Q - PubMed Abstract:
Cidofovir (1(S)-[3-hydroxy-2-(phosphonomethoxy)propyl]cytosine, CDV) is a potent inhibitor of orthopoxvirus DNA replication. Prior studies have shown that, when CDV is incorporated into a growing primer strand, it can inhibit both the 3'-to-5' exonuclease and the 5'-to-3' chain extension activities of vaccinia virus DNA polymerase. This drug can also be incorporated into DNA, creating a significant impediment to trans-lesion DNA synthesis in a manner resembling DNA damage. CDV and deoxycytidine share a common nucleobase, but CDV lacks the deoxyribose sugar. The acyclic phosphonate bears a hydroxyl moiety that is equivalent to the 3'-hydroxyl of dCMP and permits CDV incorporation into duplex DNA. To study the structural consequences of inserting CDV into DNA, we have used (1)H NMR to solve the solution structures of a dodecamer DNA duplex containing a CDV molecule at position 7 and of a control DNA duplex. The overall structures of both DNA duplexes were found to be very similar. We observed a decrease of intensity (>50%) for the imino protons neighboring the CDV (G6, T8) and the cognate base G18 and a large chemical shift change for G18. This indicates higher proton exchange rates for this region, which were confirmed using NMR-monitored melting experiments. DNA duplex melting experiments monitored by circular dichroism revealed a lower T(m) for the CDV DNA duplex (46 °C) compared to the control (58 °C) in 0.2 M salt. Our results suggest that the CDV drug is well accommodated and stable within the dodecamer DNA duplex, but the stability of the complex is less than that of the control, suggesting increased dynamics around the CDV.
Organizational Affiliation:
Department of Biochemistry, University of Alberta, 4-19 Medical Sciences Building, Edmonton, Alberta T6G 2H7, Canada.