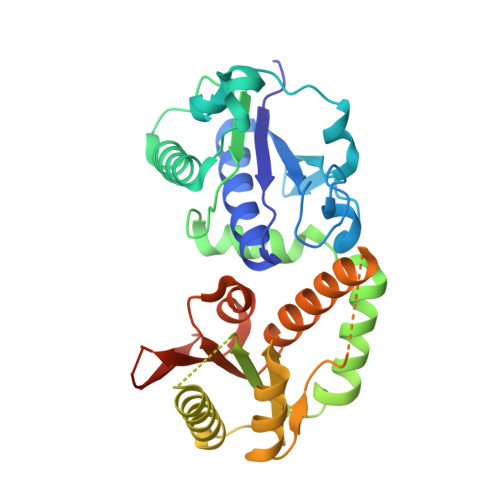
High-resolution structure of NodZ fucosyltransferase involved in the biosynthesis of the nodulation factor.
Brzezinski, K., Stepkowski, T., Panjikar, S., Bujacz, G., Jaskolski, M.(2007) Acta Biochim Pol 54: 537-549
- PubMed: 17762900
- Primary Citation of Related Structures:
2HHC, 2HLH, 2OCX - PubMed Abstract:
The fucosyltransferase NodZ is involved in the biosynthesis of the nodulation factor in nitrogen-fixing symbiotic bacteria. It catalyzes alpha1,6 transfer of l-fucose from GDP-fucose to the reducing residue of the synthesized Nod oligosaccharide. We present the structure of the NodZ protein from Bradyrhizobium expressed in Escherichia coli and crystallized in the presence of phosphate ions in two crystal forms. The enzyme is arranged into two domains of nearly equal size. Although NodZ falls in one broad class (GT-B) with other two-domain glycosyltransferases, the topology of its domains deviates from the canonical Rossmann fold, with particularly high distortions in the N-terminal domain. Mutational data combined with structural and sequence alignments indicate residues of potential importance in GDP-fucose binding or in the catalytic mechanism. They are all clustered in three conserved sequence motifs located in the C-terminal domain.
Organizational Affiliation:
Department of Crystallography, Faculty of Chemistry, A. Mickiewicz University, Poznań, Poland.