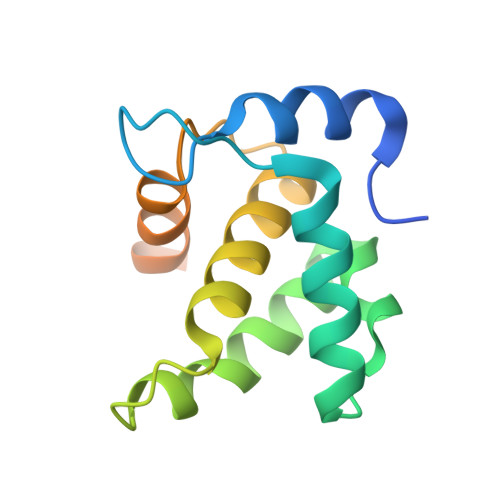
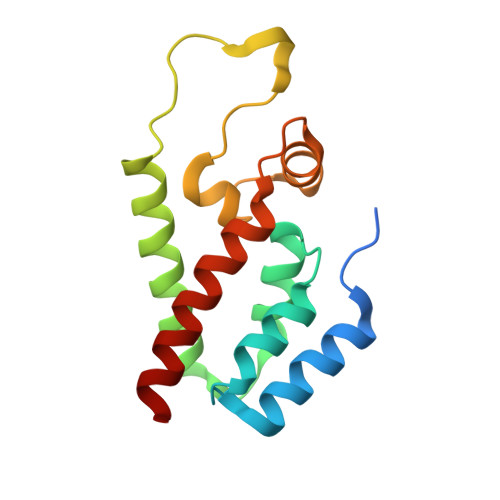
Implications for Viral Capsid Assembly from Crystal Structures of HIV-1 Gag 1-278 and CAN 133-278.
Kelly, B.N., Howard, B.R., Wang, H., Robinson, H., Sundquist, W.I., Hill, C.P.(2006) Biochemistry 45: 11257-11266
- PubMed: 16981686
- DOI: https://doi.org/10.1021/bi060927x
- Primary Citation of Related Structures:
2GOL, 2GON - PubMed Abstract:
Gag, the major structural protein of retroviruses such as HIV-1, comprises a series of domains connected by flexible linkers. These domains drive viral assembly by mediating multiple interactions between adjacent Gag molecules and by binding to viral genomic RNA and host cell membranes. Upon viral budding, Gag is processed by the viral protease to liberate distinct domains as separate proteins. The first two regions of Gag are MA, a membrane-binding module, and CA, which is a two-domain protein that makes important Gag-Gag interactions, forms the cone-shaped outer shell of the core (the capsid) in the mature HIV-1 particle, and makes an important interaction with the cellular protein cyclophilin A (CypA). Here, we report crystal structures of the mature CA N-terminal domain (CA(N)(133-278)) and a MA-CA(N) fusion (Gag(1-278)) at resolutions/R(free) values of 1.9 A/25.7% and 2.2 A/25.8%, respectively. Consistent with earlier studies, a comparison of these structures indicates that processing at the MA-CA junction causes CA to adopt an N-terminal beta-hairpin conformation that seems to be required for capsid morphology and viral infectivity. In contrast with an NMR study (Tang, C., et al. (2002) Nat. Struct. Biol. 9, 537-543), structural overlap reveals only small relative displacements for helix 6, which is located between the beta-hairpin and the CypA-binding loop. These observations argue against the proposal that CypA binding is coupled with beta-hairpin formation and support an earlier surface plasmon resonance study (Yoo, S., et al. (1997) J. Mol. Biol. 269, 780-795), which concluded that beta-hairpin formation and CypA-binding are energetically independent events.
Organizational Affiliation:
Department of Biochemistry, University of Utah, Salt Lake City, Utah 84112-5650, USA.