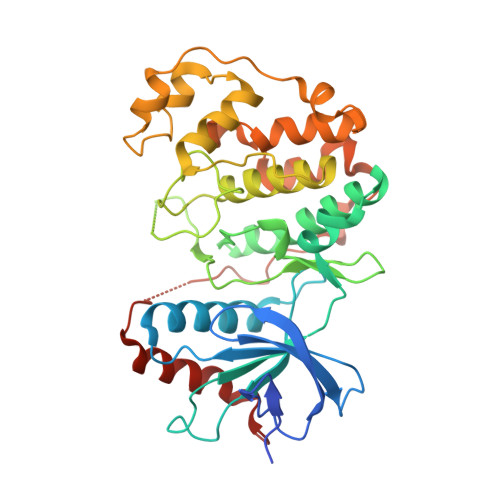
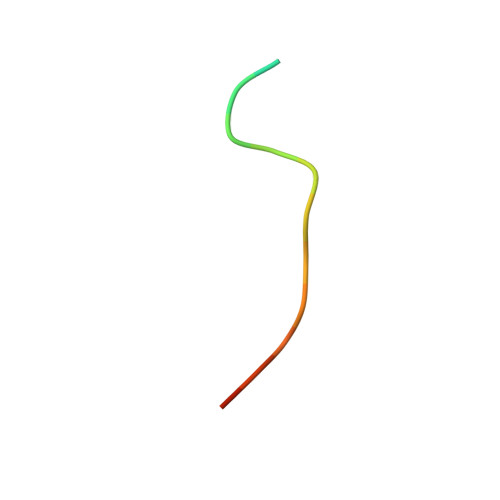
Structural basis of docking interactions between ERK2 and MAP kinase phosphatase 3
Liu, S., Sun, J.P., Zhou, B., Zhang, Z.Y.(2006) Proc Natl Acad Sci U S A 103: 5326-5331
- PubMed: 16567630
- DOI: https://doi.org/10.1073/pnas.0510506103
- Primary Citation of Related Structures:
2FYS - PubMed Abstract:
Mitogen-activated protein (MAP) kinases are central components of signal transduction pathways for cell proliferation, stress responses, and differentiation. Signaling efficiency and specificity are modulated in large part by docking interactions between individual MAP kinase and the kinase interaction motif (KIM), (R/K)(2-3)-X(1-6)-Phi(A)-X-Phi(B), in its cognate kinases, phosphatases, scaffolding proteins, and substrates. We have determined the crystal structure of extracellular signal-regulated protein kinase 2 bound to the KIM peptide from MAP kinase phosphatase 3, an extracellular signal-regulated protein kinase 2-specific phosphatase. The structure reveals that the KIM docking site, situated in a noncatalytic region opposite of the kinase catalytic pocket, is comprised of a highly acidic patch and a hydrophobic groove, which engage the basic and Phi(A)-X-Phi(B) residues, respectively, in the KIM sequence. The specific docking interactions observed in the structure consolidate all known biochemical data. In addition, structural comparison indicates that the KIM docking site is conserved in all MAP kinases. The results establish a structural model for understanding how MAP kinases interact with their regulators and substrates and provide new insights into how MAP kinase docking specificity can be achieved.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Indiana University School of Medicine, 635 Barnhill Drive, Indianapolis, IN 46202, USA.