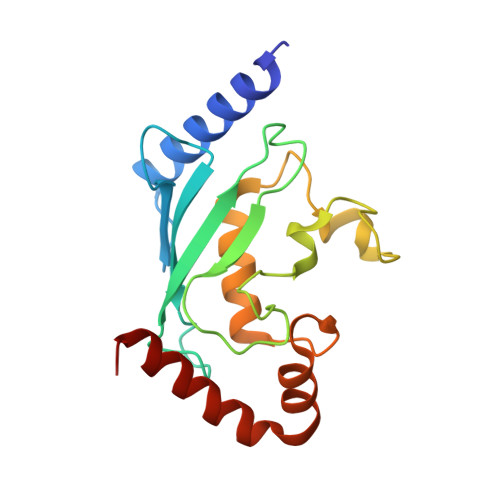
Structure of human ubiquitin-conjugating enzyme E2 G2 (UBE2G2/UBC7)
Arai, R., Yoshikawa, S., Murayama, K., Imai, Y., Takahashi, R., Shirouzu, M., Yokoyama, S.(2006) Acta Crystallogr Sect F Struct Biol Cryst Commun 62: 330-334
- PubMed: 16582478
- DOI: https://doi.org/10.1107/S1744309106009006
- Primary Citation of Related Structures:
2CYX - PubMed Abstract:
The human ubiquitin-conjugating enzyme E2 G2 (UBE2G2/UBC7) is involved in protein degradation, including a process known as endoplasmic reticulum-associated degradation (ERAD). The crystal structure of human UBE2G2/UBC7 was solved at 2.56 angstroms resolution. The UBE2G2 structure comprises a single domain consisting of an antiparallel beta-sheet with four strands, five alpha-helices and two 3(10)-helices. Structural comparison of human UBE2G2 with yeast Ubc7 indicated that the overall structures are similar except for the long loop region and the C-terminal helix. Superimposition of UBE2G2 on UbcH7 in a c-Cbl-UbcH7-ZAP70 ternary complex suggested that the two loop regions of UBE2G2 interact with the RING domain in a similar way to UbcH7. In addition, the extra loop region of UBE2G2 may interact with the RING domain or its neighbouring region and may be involved in the binding specificity and stability.
Organizational Affiliation:
Protein Research Group, RIKEN Genomic Sciences Center, Tsurumi, Yokohama 230-0045, Japan.