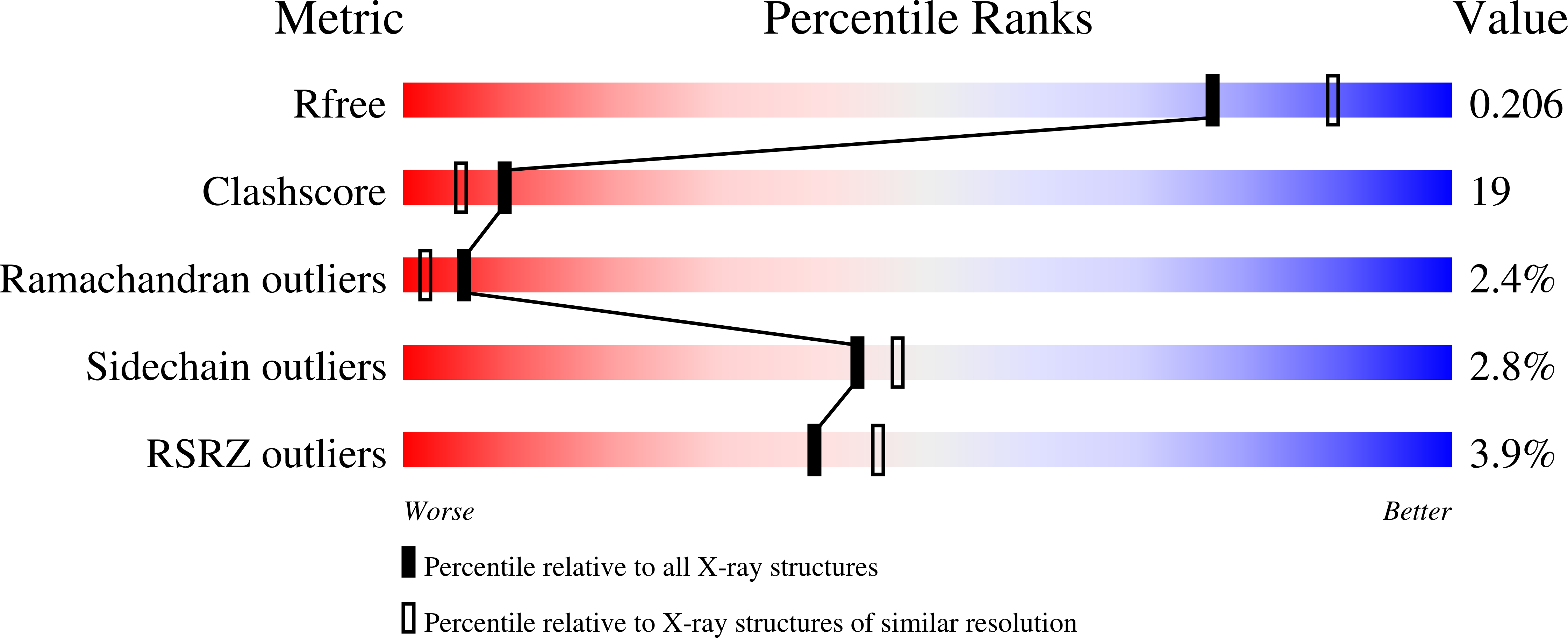
Modulator of drug activity B from Escherichia coli: crystal structure of a prokaryotic homologue of DT-diaphorase.
Adams, M.A., Jia, Z.(2006) J Mol Biol 359: 455-465
- PubMed: 16630630
- DOI: https://doi.org/10.1016/j.jmb.2006.03.053
- Primary Citation of Related Structures:
2AMJ, 2B3D - PubMed Abstract:
Modulator of drug activity B (MdaB) is a putative member of the DT-diaphorase family of NAD(P)H:oxidoreductases that afford cellular protection against quinonoid compounds. While there have been extensive investigations of mammalian homologues, putative prokaryotic members of this enzyme family have received little attention. The three-dimensional crystal structure of apo-MdaB reported herein exhibits significant structural similarity to a number of flavoproteins, including the mammalian DT-diaphorases. We have shown by mass spectrometry that the endogenously associated cofactor is flavin adenine dinucleotide and we present here the structure of MdaB in complex with this compound. Growth of Escherichia coli carrying null mutations in the genes encoding MdaB or quinol monooxygenase, the gene for which shares the mdaB promoter, were not affected by the presence of menadione. However, over-expression of recombinant quinol monooxygenase conferred a state of resistance against both tetracycline and adriamycin. This work suggests that the redox cycle formed by these proteins protects E. coli from the toxic effects of polyketide compounds rather than the oxidative stress of menadione alone.
Organizational Affiliation:
Department of Biochemistry, Queen's University, Kingston, Ont., Canada K7L 3N6.