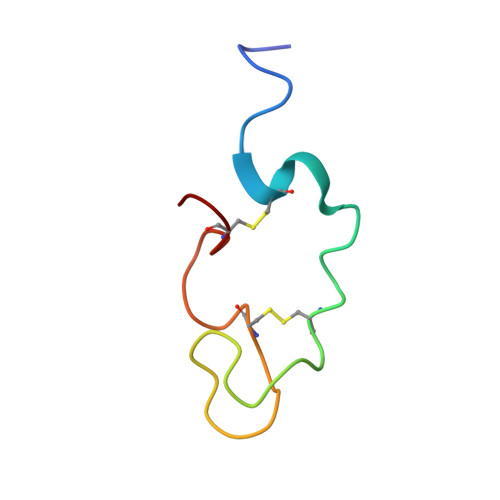
Human Lactoferricin Is Partially Folded in Aqueous Solution and Is Better Stabilized in a Membrane Mimetic Solvent
Hunter, H.N., Demcoe, A.R., Jenssen, H., Gutteberg, T.J., Vogel, H.J.(2005) Antimicrob Agents Chemother 49: 3387-3395
- PubMed: 16048952
- DOI: https://doi.org/10.1128/AAC.49.8.3387-3395.2005
- Primary Citation of Related Structures:
1Z6V, 1Z6W - PubMed Abstract:
Lactoferricins are highly basic bioactive peptides that are released in the stomach through proteolytic cleavage of various lactoferrin proteins. Here we have determined the solution structure of human lactoferricin (LfcinH) by conventional two-dimensional nuclear magnetic resonance methods in both aqueous solution and a membrane mimetic solvent. Unlike the 25-residue bovine lactoferricin (LfcinB), which adopts a somewhat distorted antiparallel beta sheet, the longer LfcinH peptide shows a helical content from Gln14 to Lys29 in the membrane mimetic solvent but a nonexistent beta-sheet character in either the N- or C-terminal regions of the peptide. The helical characteristic of the LfcinH peptide resembles the conformation that this region adopts in the crystal structure of the intact protein. The LfcinH structure determined in aqueous solution displays a nascent helix in the form of a coiled conformation in the region from Gln14 to Lys29. Numerous hydrophobic interactions create the basis for the better-defined overall structure observed in the membrane mimetic solvent. The 49-residue LfcinH peptide isolated for these studies was found to be slightly longer than previously reported peptide preparations and was found to have an intact peptide bond between residues Ala11 and Val12. The distinct solution structures of LfcinH and LfcinB represent a novel difference in the physical properties of these two peptides, which contributes to their unique physiological activities.
Organizational Affiliation:
Department of Biological Sciences, University of Calgary, 2500 University N.W., Calgary, Alberta, Canada.