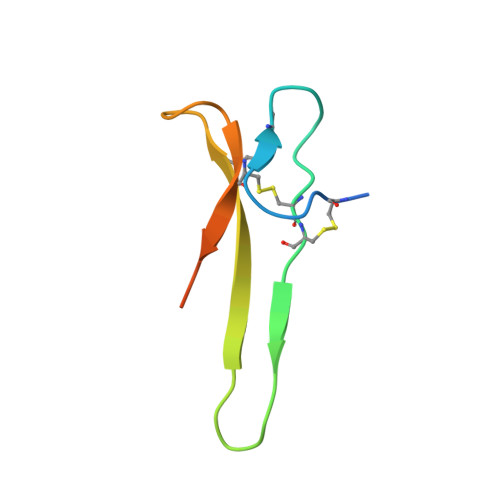
Solution structure of the carboxyl-terminal cysteine-rich domain of the VHv1.1 polydnaviral gene product: comparison with other cystine knot structural folds
Einerwold, J., Jaseja, J., Hapner, K., Webb, B., Copie, V.(2001) Biochemistry 40: 14404-14412
- PubMed: 11724552
- DOI: https://doi.org/10.1021/bi011499s
- Primary Citation of Related Structures:
1XI7, 1XJ1 - PubMed Abstract:
Polydnaviruses are an unusual group of insect viruses that have an obligate symbiotic association with certain parasitic wasps. These viruses are transmitted with the wasp egg during oviposition into lepidopteran insects, enabling the survival and development of the egg inside the host larvae. We report the three-dimensional structure of a novel polydnaviral cysteine-rich motif (cys-motif), identified as the carboxyl-terminal domain of a two cys-motif containing polydnaviral VHv1.1 gene product, abbreviated "C-term VHv1.1". This 65-residue domain was identified experimentally by limited proteolysis of the full-length protein and was subsequently cloned in a bacterial expression system for NMR studies. The C-term VHv1.1 3D structure was determined in solution by two-dimensional (1)H NMR spectroscopy. Calculation of the structure was based on a total of 300 upper distance restraints and 20 dihedral angle constraints, and resulted in an ensemble of 25 representative conformers with an average rmsd of 0.47 A from the mean structure for core backbone atoms. The protein core is made of a four beta-strand scaffold held together in a compact structure by three disulfide bonds, which form a cystine knot. The four beta-strands are arranged in an unusual configuration to form a triple-stranded beta-sheet and double-stranded beta-sheet. Comparison with other classes of cystine knots provides indication that C-term VHv1.1 represents a new and distinct cystine knot motif. This analysis provides a structural basis for interpretation of the genetic and amino acid sequence data classifying polydnavirus gene products as members of cysteine-rich protein families.
Organizational Affiliation:
Department of Chemistry and Biochemistry, Montana State University, 108 Gaines Hall, Bozeman, Montana 59717, USA.