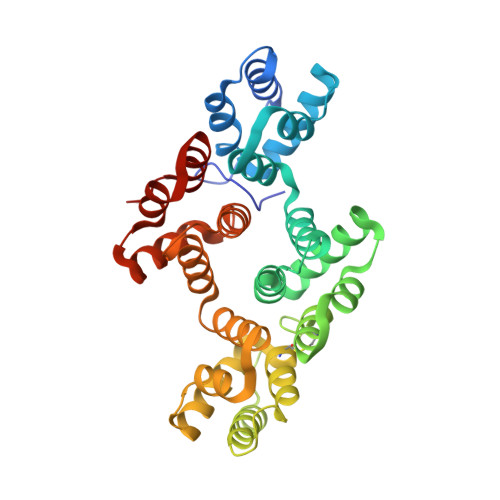
The Crystal Structure of Annexin A8 is Similar to that of Annexin A3
Rety, S., Sopkova-De Oliveira Santos, J., Dreyfuss, L., Blondeau, K., Hofbauerova, K., Raguenes-Nicol, C., Kerboeuf, D., Renouard, M., Russo-Marie, F., Lewit-Bentley, A.(2005) J Mol Biol 345: 1131
- PubMed: 15644210
- DOI: https://doi.org/10.1016/j.jmb.2004.11.015
- Primary Citation of Related Structures:
1W3W, 1W45 - PubMed Abstract:
Annexin A8 is a relatively infrequent and poorly studied member of this large family of calcium-binding and membrane-binding proteins. It is, however, associated with a specific disease, acute promyelocytic leukemia. We have solved its three-dimensional structure, which includes a moderately long and intact N terminus. The structure is closest to that of annexin A3 and highlights several important regions of inherent flexibility in the annexin molecule. The N terminus resembles that of annexin A3, as it lies along the concave surface of the molecule and inserts partially into the hydrophilic channel in its centre. Since both annexins A3 and A8 are expressed in promyelocytic cells during their differentiation, the similarity in their structures might suggest a functional relationship.
Organizational Affiliation:
LURE, Centre Universitaire Paris-Sud, BP 34, 91898 Orsay Cedex, France.