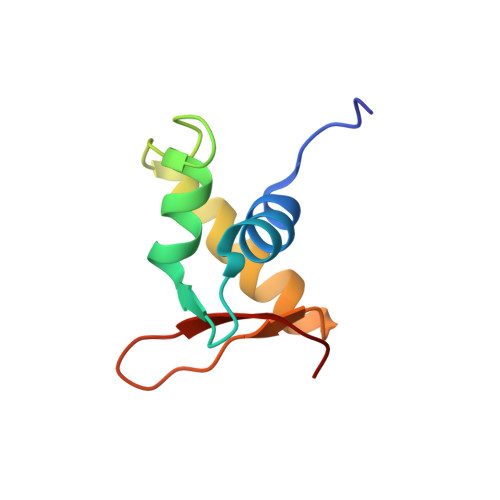
The linker histone homolog Hho1p from Saccharomyces cerevisiae represents a winged helix-turn-helix fold as determined by NMR spectroscopy.
Ono, K., Kusano, O., Shimotakahara, S., Shimizu, M., Yamazaki, T., Shindo, H.(2003) Nucleic Acids Res 31: 7199-7207
- PubMed: 14654695
- DOI: https://doi.org/10.1093/nar/gkg931
- Primary Citation of Related Structures:
1UHM - PubMed Abstract:
Hho1p is assumed to serve as a linker histone in Saccharomyces cerevisiae and, notably, it possesses two putative globular domains, designated HD1 (residues 41-118) and HD2 (residues 171-252), that are homologous to histone H5 from chicken erythrocytes. We have determined the three-dimensional structure of globular domain HD1 with high precision by heteronuclear magnetic resonance spectroscopy. The structure had a winged helix-turn-helix motif composed of an alphabetaalphaalphabetabeta fold and closely resembled the structure of the globular domain of histone H5. Interestingly, the second globular domain, HD2, in Hho1p was unstructured under physiological conditions. Gel mobility assay demonstrated that Hho1p preferentially binds to supercoiled DNA over linearized DNA. Furthermore, NMR analysis of the complex of a deletion mutant protein (residues 1-118) of Hho1p with a linear DNA duplex revealed that four regions within the globular domain HD1 are involved in the DNA binding. The above results suggested that Hho1p possesses properties similar to those of linker histones in higher eukaryotes in terms of the structure and binding preference towards supercoiled DNA.
Organizational Affiliation:
School of Life Science, Tokyo University of Pharmacy and Life Science, 1432-1 Horinouchi, Hachioji, Tokyo 192-0392, Japan.