Solution Structure of the Escherichia coli YojN Histidine-phosphotransferase Domain and its Interaction with Cognate Phosphoryl Receiver Domains
Rogov, V.V., Bernhard, F., Loehr, F., Doetsch, V.(2004) J Mol Biol 343: 1035-1048
- PubMed: 15476819
- DOI: https://doi.org/10.1016/j.jmb.2004.08.096
- Primary Citation of Related Structures:
1SR2 - PubMed Abstract:
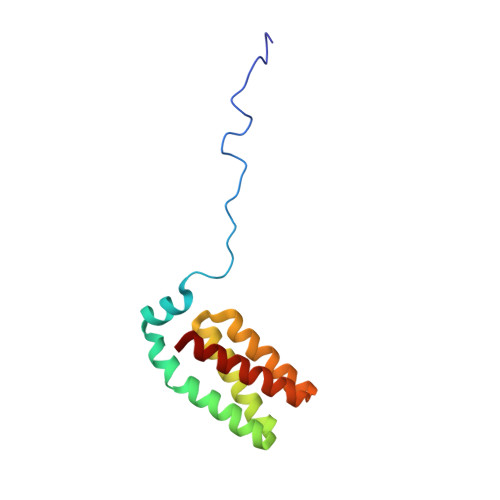
The Rcs signaling system in Escherichia coli controls a variety of physiological functions, including capsule synthesis, cell division and motility. The activity of the central regulator RcsB is modulated by phosphorylation through the sensor kinases YojN and RcsC, with the YojN histidine phosphotransferase (HPt) domain representing the catalytic unit that coordinates the potentially reversible phosphotransfer reaction between the receiver domains of the RcsB and RcsC proteins. Heteronuclear high-resolution NMR spectroscopy was employed to determine the solution structure of the YojN-HPt domain and to map the interaction with its two cognate receiver domains. The solution structure of YojN-HPt exhibits a well-ordered and rigid protein core consisting of the five helices alphaI to alphaV. The helices alphaII to alphaV form a four-helix bundle signature motif common to proteins of similar function, and helix alphaI forms a cap on top of the bundle. The helix alphaII is separated by a proline induced kink into two parts with different orientations and dynamic behavior that is potentially important for complex formation with other proteins. The N-terminal part of YojN-HPt spanning the first 26 amino acid residues seems to contain neither a regular secondary structure nor a stable tertiary structure and is disordered in solution. The identified YojN-HPt recognition sites for the regulator RcsB and for the isolated receiver domain of the RcsC kinase largely overlap in defined regions of the helices alphaII and alphaIII, but show significant differences. Using the residues with the largest chemical shift changes obtained from titration experiments, we observed a dissociation constant of approximately 200microM for YojN-HPt/RcsC-PR and of 40microM for YojN-HPt/RcsB complexes. Our data indicate the presence of a recognition area in close vicinity to the active-site histidine residue of HPt domains as a determinant of specificity in signal-transduction pathways.
Organizational Affiliation:
Institute of Biophysical Chemistry, Centre for Biomolecular Magnetic Resonance, J.W. Goethe-University, Frankfurt-am-Main, Germany.