From structure and dynamics of protein L7/L12 to molecular switching in ribosome.
Bocharov, E.V., Sobol, A.G., Pavlov, K.V., Korzhnev, D.M., Jaravine, V.A., Gudkov, A.T., Arseniev, A.S.(2004) J Biol Chem 279: 17697-17706
- PubMed: 14960595
- DOI: https://doi.org/10.1074/jbc.M313384200
- Primary Citation of Related Structures:
1RQS, 1RQT, 1RQU, 1RQV - PubMed Abstract:
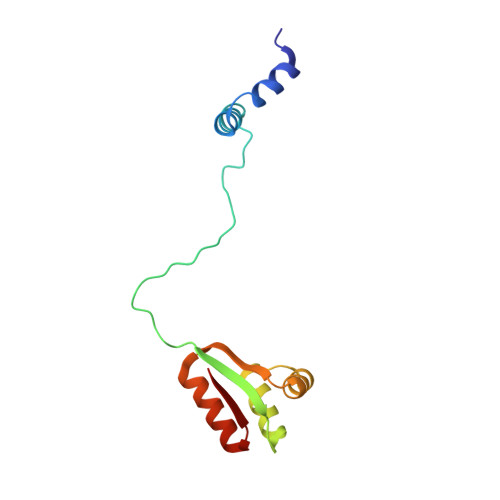
Based on the (1)H-(15)N NMR spectroscopy data, the three-dimensional structure and internal dynamic properties of ribosomal protein L7 from Escherichia coli were derived. The structure of L7 dimer in solution can be described as a set of three distinct domains, tumbling rather independently and linked via flexible hinge regions. The dimeric N-terminal domain (residues 1-32) consists of two antiparallel alpha-alpha-hairpins forming a symmetrical four-helical bundle, whereas the two identical C-terminal domains (residues 52-120) adopt a compact alpha/beta-fold. There is an indirect evidence of the existence of transitory helical structures at least in the first part (residues 33-43) of the hinge region. Combining structural data for the ribosomal protein L7/L12 from NMR spectroscopy and x-ray crystallography, it was suggested that its hinge region acts as a molecular switch, initiating "ratchet-like" motions of the L7/L12 stalk with respect to the ribosomal surface in response to elongation factor binding and GTP hydrolysis. This hypothesis allows an explanation of events observed during the translation cycle and provides useful insights into the role of protein L7/L12 in the functioning of the ribosome.
Organizational Affiliation:
Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, ul. Miklukho-Maklaya, 16/10, Moscow 117997, Russia.