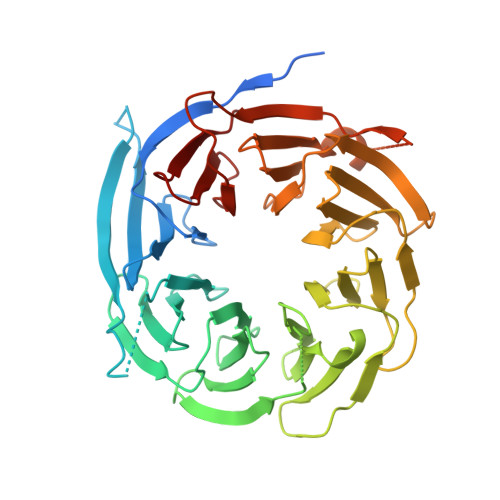
The structure of Sif2p, a WD repeat protein functioning in the SET3 corepressor complex.
Cerna, D., Wilson, D.K.(2005) J Mol Biol 351: 923-935
- PubMed: 16051270
- DOI: https://doi.org/10.1016/j.jmb.2005.06.025
- Primary Citation of Related Structures:
1R5M - PubMed Abstract:
In Saccharomyces cerevisiae, the SIF2 gene product is an integral component of the Set3 complex (SET3C), an assembly of proteins with some homology to the human SMRT and N-CoR corepressor complexes. SET3C has histone deacetylase activity that is responsible for repressing a set of meiotic genes. We have determined the X-ray crystal structure of a 46 kDa C-terminal domain of a SET3C core protein, Sif2p to 1.55 A resolution and a crystallographic R-factor of 19.0%. This domain contains an unusual eight-bladed beta-propeller structure, which differs from other transcriptional corepressor structures such as yeast Tup1p and human groucho (Gro)/TLE1, which have only seven. We have demonstrated intact Sif2p is a tetramer and the N-terminal LisH (Lis-homology)-containing domain mediates tetramerization and interaction with another component of SET3C, Snt1p. Multiple sequence alignments indicate that a surface on the "top" of the protein is conserved among species, suggesting that it may play a common role in binding partner proteins. Since Sif2p appears to be the yeast homolog of human TBL1 and TBLR1, which function in the N-CoR/SMRT complexes, its structural and oligomeric properties are likely to be very similar.
Organizational Affiliation:
Section of Molecular and Cellular Biology, University of California, Davis, CA 95616, USA.