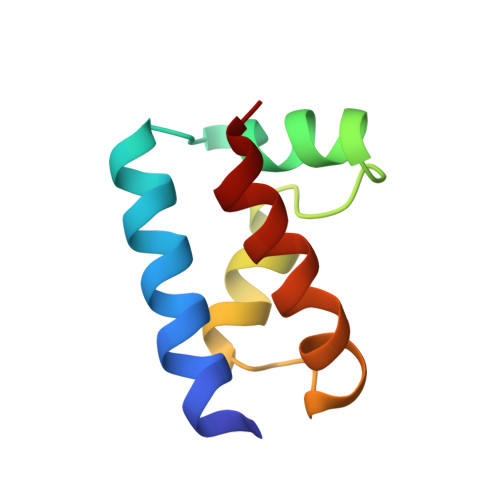
The solution structure of the Oct-1 POU-specific domain reveals a striking similarity to the bacteriophage lambda repressor DNA-binding domain.
Assa-Munt, N., Mortishire-Smith, R.J., Aurora, R., Herr, W., Wright, P.E.(1993) Cell 73: 193-205
- PubMed: 8462099
- DOI: https://doi.org/10.1016/0092-8674(93)90171-l
- Primary Citation of Related Structures:
1POU - PubMed Abstract:
The POU-specific (POUs) domain, in association with a POU-type homeodomain, forms the bipartite DNA-binding POU domain. The solution structure of the Oct-1 POUs domain has been determined by multidimensional nuclear magnetic resonance spectroscopy and consists of four alpha helices surrounding a conserved hydrophobic core. The POUs domain is structurally similar to the DNA-binding domains of the bacteriophage lambda and 434 repressors and 434 Cro. These domains exhibit superimposable helix-turn-helix (HTH) motifs, except that in the POUs domain, the first helix and the linker to the second helix of the motif are extended. The conserved structural features have been used to propose a plausible model for DNA binding by the POUs domain. A human dwarfism mutation that affects positive control in the related POU domain protein Pit-1 maps to the same region of the HTH motif as do positive control mutations in lambda repressor.
Organizational Affiliation:
Department of Molecular Biology, Scripps Research Institute, La Jolla, California 92037.