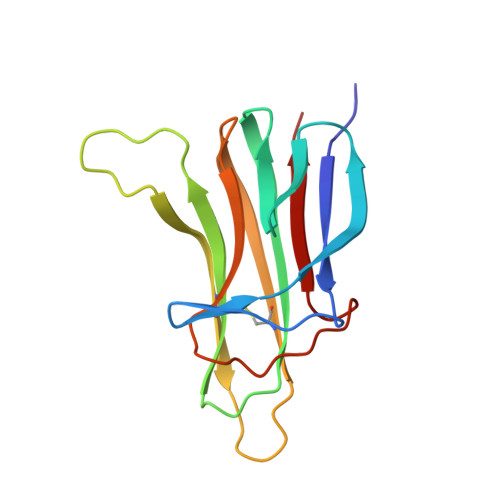
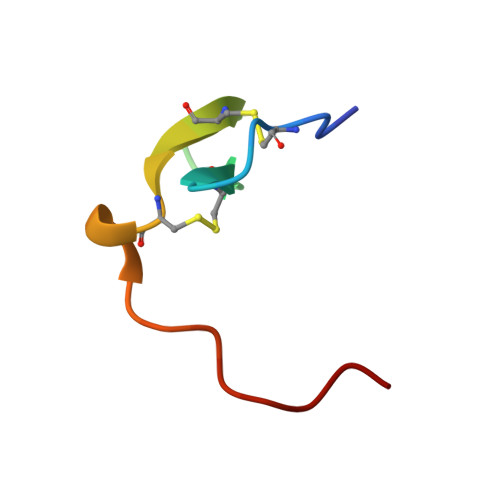
Ligand-receptor binding revealed by the TNF family member TALL-1.
Liu, Y., Hong, X., Kappler, J., Jiang, L., Zhang, R., Xu, L., Pan, C.H., Martin, W.E., Murphy, R.C., Shu, H.B., Dai, S., Zhang, G.(2003) Nature 423: 49-56
- PubMed: 12721620
- DOI: https://doi.org/10.1038/nature01543
- Primary Citation of Related Structures:
1OQD, 1OQE - PubMed Abstract:
The tumour necrosis factor (TNF) ligand TALL-1 and its cognate receptors, BCMA, TACI and BAFF-R, were recently identified as members of the TNF superfamily, which are essential factors contributing to B-cell maturation. The functional, soluble fragment of TALL-1 (sTALL-1) forms a virus-like assembly for its proper function. Here we determine the crystal structures of sTALL-1 complexed with the extracellular domains of BCMA and BAFF-R at 2.6 and 2.5 A, respectively. The single cysteine-rich domain of BCMA and BAFF-R both have saddle-like architectures, which sit on the horseback-like surface formed by four coil regions on each individual sTALL-1 monomer. Three novel structural modules, D2, X2 and N, were revealed from the current structures. Sequence alignments, structural modelling and mutagenesis revealed that one disulphide bridge in BAFF-R is critical for determining the binding specificity of the extracellular domain eBAFF-R to TALL-1 instead of APRIL, a closely related ligand of TALL-1, which was confirmed by binding experiments in vitro.
Organizational Affiliation:
Integrated Department of Immunology, National Jewish Medical and Research Center, Denver, Colorado 80206, USA.