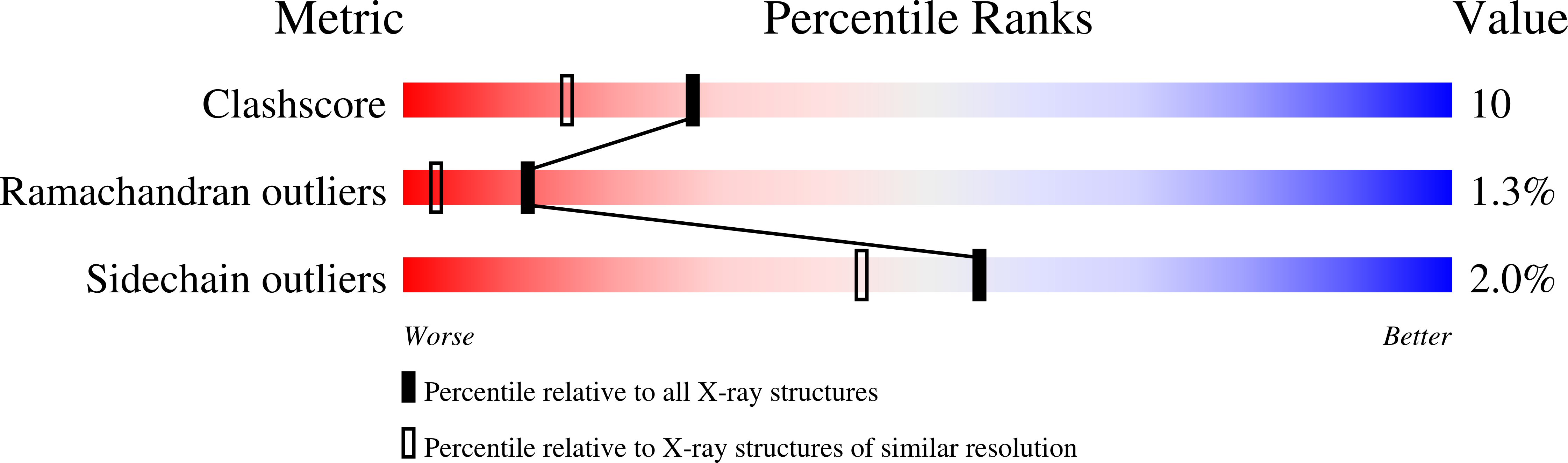Complex of rat transthyretin with tetraiodothyroacetic acid refined at 2.1 and 1.8 A resolution.
Muziol, T., Cody, V., Luft, J.R., Pangborn, W., Wojtczak, A.(2001) Acta Biochim Pol 48: 877-884
- PubMed: 11995998
- Primary Citation of Related Structures:
1KGI - PubMed Abstract:
The crystal structure of rat transthyretin (rTTR) complex with 3,5,3',5'-tetraiodothyroacetic acid (T4Ac) was determined at 1.8 A resolution with low temperature synchrotron data collected at CHESS. The structure was refined to R = 0.207 and Rfree = 0.24 with the use of 8-1.8 A data. The additional 8000 reflections from the incomplete 2.1-1.8 data shell, included in the refinement, reduced the Rfree index by 1.3%. Structure comparison with the model refined against the complete 8-2.1 A data revealed no differences in the ligand orientation and the conformation of the polypeptide chain in the core regions. However, the high-resolution data included in the refinement improved the model in the flexible regions poorly defined with the lower resolution data. Also additional sixteen water molecules were found in the difference map calculated with the extended data. The structure revealed both forward and reverse binding of tetraiodothyroacetic acid in one binding site and two modes of forward ligand binding in the second site, with the phenolic iodine atoms occupying different sets of the halogen binding pockets.
Organizational Affiliation:
Institute of Chemistry, Nicolaus Copernicus University, Toruń, Poland.