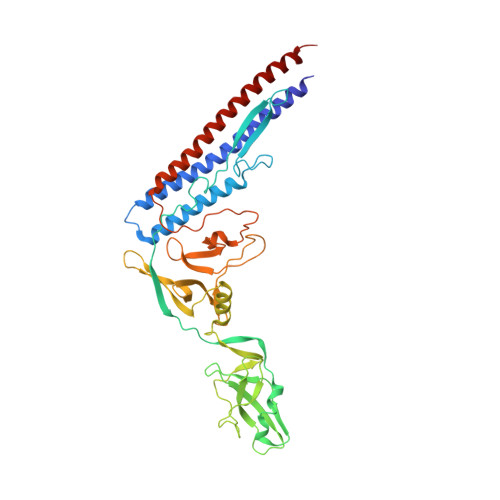
Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling
Samatey, F.A., Imada, K., Nagashima, S., Vondervisz, F., Kumasaka, T., Yamamoto, M., Namba, K.(2001) Nature 410: 331-337
- PubMed: 11268201
- DOI: https://doi.org/10.1038/35066504
- Primary Citation of Related Structures:
1IO1 - PubMed Abstract:
The bacterial flagellar filament is a helical propeller constructed from 11 protofilaments of a single protein, flagellin. The filament switches between left- and right-handed supercoiled forms when bacteria switch their swimming mode between running and tumbling. Supercoiling is produced by two different packing interactions of flagellin called L and R. In switching from L to R, the intersubunit distance ( approximately 52 A) along the protofilament decreases by 0.8 A. Changes in the number of L and R protofilaments govern supercoiling of the filament. Here we report the 2.0 A resolution crystal structure of a Salmonella flagellin fragment of relative molecular mass 41,300. The crystal contains pairs of antiparallel straight protofilaments with the R-type repeat. By simulated extension of the protofilament model, we have identified possible switch regions responsible for the bi-stable mechanical switch that generates the 0.8 A difference in repeat distance.
Organizational Affiliation:
Protonic NanoMachine Project, ERATO, JST, 3-4 Hikaridai, Seika, Kyoto 619-0237, Japan.