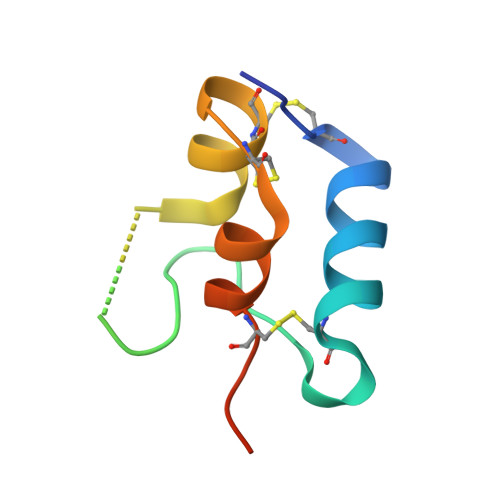
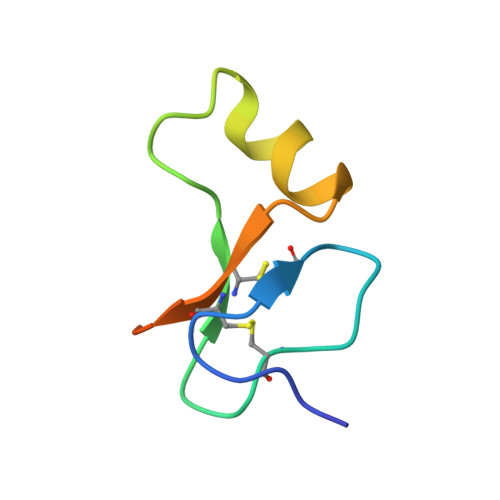
The Interaction of Insulin-Like Growth Factor-I with the N-Terminal Domain of Igfbp-5
Zesaawski, W., Beisel, H.G., Kamionka, M., Kalus, W., Engh, R.A., Huber, R., Lang, K., Holak, T.A.(2001) EMBO J 20: 3638
- PubMed: 11447105
- DOI: https://doi.org/10.1093/emboj/20.14.3638
- Primary Citation of Related Structures:
1H59 - PubMed Abstract:
Insulin-like growth factors (IGFs) are key regulators of cell proliferation, differentiation and transformation, and are thus pivotal in cancer, especially breast, prostate and colon neoplasms. They are also important in many neurological and bone disorders. Their potent mitogenic and anti-apoptotic actions depend primarily on their availability to bind to the cell surface IGF-I receptor. In circulation and interstitial fluids, IGFs are largely unavailable as they are tightly associated with IGF-binding proteins (IGFBPs) and are released after IGFBP proteolysis. Here we report the 2.1 A crystal structure of the complex of IGF-I bound to the N-terminal IGF-binding domain of IGFBP-5 (mini-IGFBP-5), a prototype interaction for all N-terminal domains of the IGFBP family. The principal interactions in the complex comprise interlaced hydrophobic side chains that protrude from both IGF-I and the IGFBP-5 fragment and a surrounding network of polar interactions. A solvent-exposed hydrophobic patch is located on the IGF-I pole opposite to the mini-IGFBP-5 binding region and marks the IGF-I receptor binding site.
Organizational Affiliation:
Max Planck Institute for Biochemistry, D-82152 Martinsried, Germany.