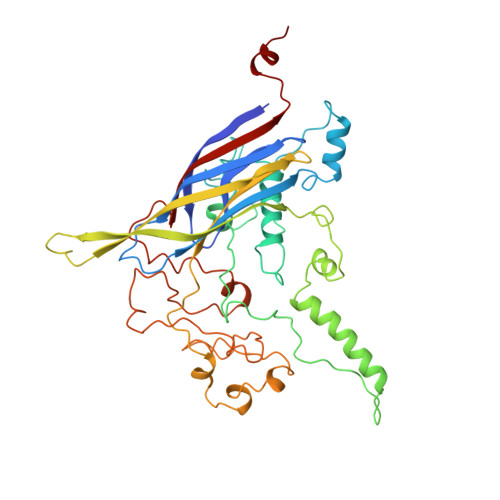
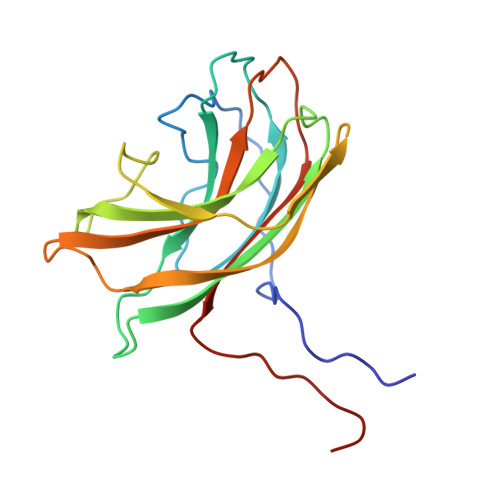
Atomic structure of the degraded procapsid particle of the bacteriophage G4: induced structural changes in the presence of calcium ions and functional implications.
McKenna, R., Bowman, B.R., Ilag, L.L., Rossmann, M.G., Fane, B.A.(1996) J Mol Biol 256: 736-750
- PubMed: 8642594
- DOI: https://doi.org/10.1006/jmbi.1996.0121
- Primary Citation of Related Structures:
1GFF - PubMed Abstract:
Bacteriophage G4 and phiX174 are members of the Microviridae family. The degree of similarity of the structural proteins ranges from 66% identity of the F protein to 40% identity of the G protein. The atomic structure of the phiX174 virion had previously been determined by X-ray crystallography. Bacteriophage G4 procapsids, consisting of the structural proteins F, G, D, B, H, and small traces of J but no DNA, were set up for crystallization. However, the resultant crystals were of degraded procapsid particles, which had lost the assembly scaffolding proteins D and B, resulting in particles that resembled empty virions. The structure of the degraded G4 procapsid has been determined to 3.0 angstrom resolution. The particles crystallized in the hexagonal space group P6(3)22 with unit cell dimensions a=b=414.2(5) angstrom and c=263.0(3) angstrom. The diffraction data were collected at the Cornell High Energy Synchrotron Source (CHESS) on film and image plates using oscillation photography. Packing considerations indicated there were two particles per unit cell. A self-rotation function confirmed that the particles were positioned on 32 point group special positions in the unit cell. Initial phases were calculated to 6 angstrom resolution, based on the known phiX174 virion model. Phase information was then extended in steps to 3.0 angstrom resolution by molecular replacement electron density modification and particle envelope generation. The resulting electron density map was readily interpretable in terms of the F and G polypeptides, as occur in the mature capsid of phiX174. In a few regions of the electron density map there were inconsistencies between the density and the published amino acid sequence. Redetermining the amino acid sequence confirmed that the density was correct. The r.m.s. deviation between the Calpha backbone of the mature capsid of phiX174 and the degraded G4 procapsid was 0.36 angstrom for the F protein and 1.38 angstrom for the G protein. This is consistent with the greater conservation of the F protein compared to the G protein sequences among members of the Microviridae family. Functionally important features between phiX174 and G4 had greater conservation. Calcium ions (Ca2+) were shown to bind to G4 at a general site located near the icosahedral 3-fold axis on the F protein capsid, equivalent to sites found previously in phiX174. Binding of Ca2+ also caused the ordering of the conserved region of the DNA binding protein J, which was present in the degraded procapsid particle in the absence of DNA.
Organizational Affiliation:
Department of Biological Sciences, Purdue University, West Lafayette, Indiana 47907 USA.