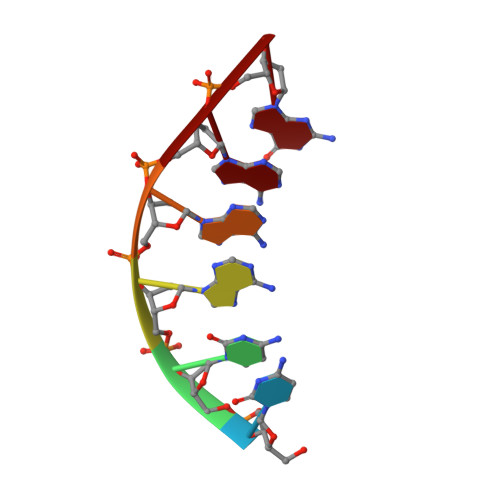
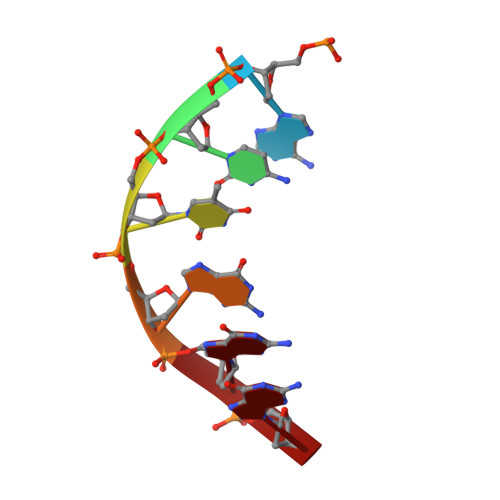
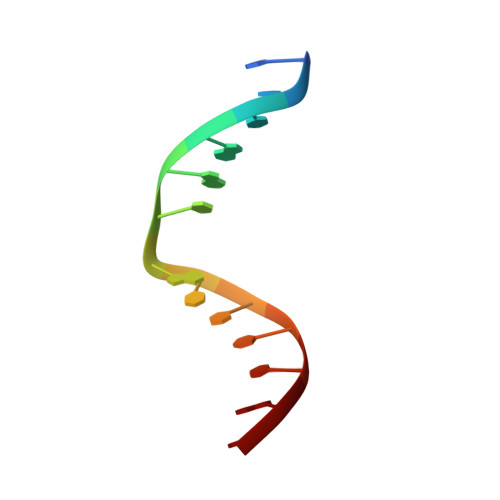
Solution structure of Co(III)-bleomycin-OOH bound to a phosphoglycolate lesion containing oligonucleotide: implications for bleomycin-induced double-strand DNA cleavage.
Hoehn, S.T., Junker, H.D., Bunt, R.C., Turner, C.J., Stubbe, J.(2001) Biochemistry 40: 5894-5905
- PubMed: 11352724
- DOI: https://doi.org/10.1021/bi002635g
- Primary Citation of Related Structures:
1G5L, 1GJ2 - PubMed Abstract:
Bleomycin (BLM) is an antitumor antibiotic that is used clinically. Its major cause of cytotoxicity is thought to be related to BLM's ability to cause double-strand (ds) DNA cleavage. A single molecule of BLM appears to cleave both strands of DNA in the presence of its required cofactors Fe(2+) and oxygen without dissociating from the helix. A mechanism for this process has been proposed based on a model structure of the hydroperoxide of Co(III)-BLM (CoBLM) bound sequence-specifically to an intact duplex containing a GTAC site, a hot spot for ds cleavage [Vanderwall, D. E., Lui, S. M., Wu, W., Turner, C. J., Kozarich, J. W., and Stubbe, J. (1997) Chem. Biol. 4, 373-387]. In this paper, we present a structural model for the second cleavage event. Two-dimensional NMR spectroscopy and molecular modeling were carried out to study CoBLM bound to d(CCAAAGXACTGGG).d(CCCAGTACTTTGG), where X represents a 3'-phosphoglycolate lesion next to a 5'-phosphate. Assignments of 729 NOEs, including 51 between the drug and the DNA and 126 within the BLM molecule, have been made. These NOEs in addition to 96 dihedral angle constraints have been used to obtain a well-defined structural model for this complex. The model reveals that the bithiazole tail is partially intercalated between the T19 and the A20 of the duplex and that the metal binding domain is poised for abstraction of the T19 H4' in the minor groove. The modeling further reveals that the predominant conformation of the bithiazole protons is trans. Two cis conformations of these protons are also observed, and ROESY experiments provide evidence for interconversion of all of these forms. The relationship of these observations to the model for ds cleavage is presented.
Organizational Affiliation:
Department of Chemistry, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, Massachusetts 02139, USA.