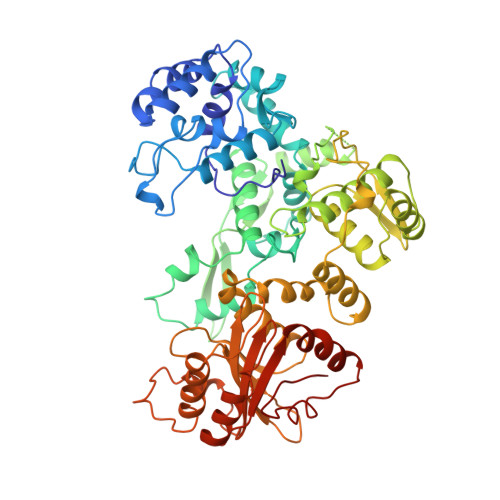
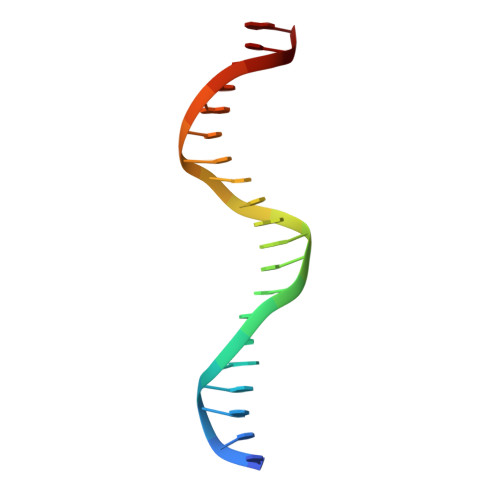
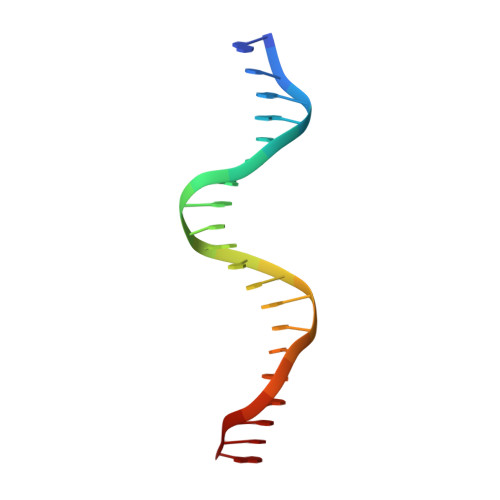
Structure of the multimodular endonuclease FokI bound to DNA.
Wah, D.A., Hirsch, J.A., Dorner, L.F., Schildkraut, I., Aggarwal, A.K.(1997) Nature 388: 97-100
- PubMed: 9214510
- DOI: https://doi.org/10.1038/40446
- Primary Citation of Related Structures:
1FOK - PubMed Abstract:
FokI is a member of an unusual class of bipartite restriction enzymes that recognize a specific DNA sequence and cleave DNA nonspecifically a short distance away from that sequence. Because of its unusual bipartite nature, FokI has been used to create artificial enzymes with new specificities. We have determined the crystal structure at 2.8A resolution of the complete FokI enzyme bound to DNA. As anticipated, the enzyme contains amino- and carboxy-terminal domains corresponding to the DNA-recognition and cleavage functions, respectively. The recognition domain is made of three smaller subdomains (D1, D2 and D3) which are evolutionarily related to the helix-turn-helix-containing DNA-binding domain of the catabolite gene activator protein CAP. The CAP core has been extensively embellished in the first two subdomains, whereas in the third subdomain it has been co-opted for protein-protein interactions. Surprisingly, the cleavage domain contains only a single catalytic centre, raising the question of how monomeric FokI manages to cleave both DNA strands. Unexpectedly, the cleavage domain is sequestered in a 'piggyback' fashion by the recognition domain. The structure suggests a new mechanism for nuclease activation and provides a framework for the design of chimaeric enzymes with altered specificities.
Organizational Affiliation:
Department of Biochemistry and Molecular Biophysics, Columbia University, New York 10032, USA.