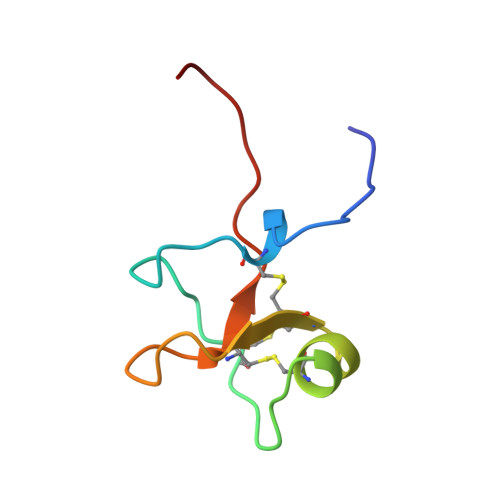
High-Resolution Solution Structure of Human Intestinal Trefoil Factor and Functional Insights from Detailed Structural Comparisons with the Other Members of the Trefoil Family of Mammalian Cell Motility Factors
Lemercinier, X., Muskett, F.W., Cheeseman, B., Mcintosh, P.B., Thim, L., Carr, M.D.(2001) Biochemistry 40: 9552
- PubMed: 11583154
- DOI: https://doi.org/10.1021/bi010184+
- Primary Citation of Related Structures:
1E9T - PubMed Abstract:
The secreted proteins intestinal trefoil factor (ITF, 59 residues), pS2 (60 residues), and spasmolytic polypeptide (SP, 106 residues) form a small family of trefoil domain-containing mammalian cell motility factors, which are essential for the maintenance of all mucous-coated epithelial surfaces. We have used 1H NMR spectroscopy to determine the high-resolution structure of human ITF, which has allowed detailed structural comparisons with the other trefoil cell motility factors. The conformation of residues 10-53 of hITF is determined to high precision, but the structure of the N- and C-terrminal residues is poorly defined by the NMR data, which is probably indicative of significant mobility. The core of the trefoil domain in hITF consists of a two-stranded antiparallel beta-sheet (Cys 36 to Asp 39 and Trp 47 to Lys 50), which is capped by an irregular loop and forms a central hairpin (loop 3). The beta-sheet is preceded by a short alpha-helix (Lys 29 to Arg 34), with the majority of the remainder of the domain contained in two loops formed from His 25 to Pro 28 (loop 2) and Ala 12 to Arg 18 (loop 1), which lie on either side of the central hairpin. The region formed by the surface of loop 2, the cleft between loop 2 and loop 3, and the adjacent face of loop 3 has previously been proposed to form the functional site of trefoil domains. Detailed comparisons of the backbone conformations and surface features of the family of trefoil cell motility factors (porcine SP, pS2, and hITF) have identified significant structural and electrostatic differences in the loop 2/loop 3 regions, which suggest that each trefoil protein has a specific target or group of target molecules.
Organizational Affiliation:
Laboratory of Molecular Structure, National Institute for Biological Standards and Control, Potters Bar, Hertfordshire, UK.