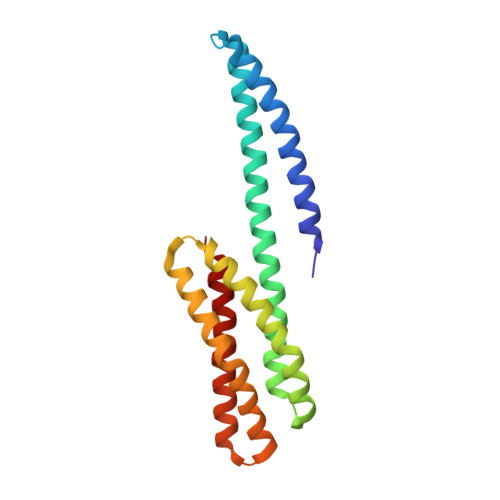
Structure of the dimerization and beta-catenin-binding region of alpha-catenin.
Pokutta, S., Weis, W.I.(2000) Mol Cell 5: 533-543
- PubMed: 10882138
- DOI: https://doi.org/10.1016/s1097-2765(00)80447-5
- Primary Citation of Related Structures:
1DOV, 1DOW - PubMed Abstract:
In adherens junctions, alpha-catenin links the cadherin-beta-catenin complex to the actin-based cytoskeleton. alpha-catenin is a homodimer in solution, but forms a 1:1 heterodimer with beta-catenin. The crystal structure of the alpha-catenin dimerization domain, residues 82-279, shows that alpha-catenin dimerizes through formation of a four-helix bundle in which two antiparallel helices are contributed by each protomer. A slightly larger fragment, comprising residues 57-264, binds to beta-catenin. A chimera consisting of the alpha-catenin-binding region of beta-catenin linked to the amino terminus of alpha-catenin 57-264 behaves as a monomer in solution, as expected, since beta-catenin binding disrupts the alpha-catenin dimer. The crystal structure of this chimera reveals the interaction between alpha- and beta-catenin, and provides a basis for understanding adherens junction assembly.
Organizational Affiliation:
Department of Structural Biology, Stanford University School of Medicine, California 94305, USA.