A phylogenetic approach to target selection for structural genomics: solution structure of YciH.
Cort, J.R., Koonin, E.V., Bash, P.A., Kennedy, M.A.(1999) Nucleic Acids Res 27: 4018-4027
- PubMed: 10497266
- DOI: https://doi.org/10.1093/nar/27.20.4018
- Primary Citation of Related Structures:
1D1R - PubMed Abstract:
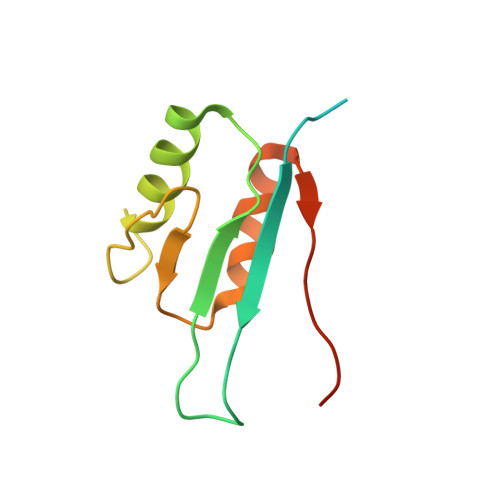
Structural genomics presents an enormous challenge with up to 100 000 protein targets in the human genome alone. At current rates of structure deter-mination, judicious selection of targets is necessary. Here, a phylogenetic approach to target selection is described which makes use of the National Center for Biotechnology Information database of Clusters of Orthologous Groups (COGS). The strategy is designed so that each new protein structure is likely to provide novel sequence-fold information. To demonstrate this approach, the NMR solution structure of YciH (COG0023), a putative translation initiation factor from Escherichia coli, has been determined and its fold classified. YciH is an ortholog of eIF-1/SUI1, an integral component of the translation initiation complex in eukaryotes. The structure consists of two antiparallel alpha-helices packed against the same side of a five-stranded beta-sheet. The first 31 residues of the 11.5 kDa protein are unstructured in solution. Comparative analysis indicates that the folded portion of YciH resembles a number of structures with the alpha-beta plait topology, though its sequence is not homologous to any of them. Thus, the phylogenetic approach to target selection described here was used successfully to identify a new homologous superfamily within this topology.
Organizational Affiliation:
Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory, Richland, WA 99352, USA.