Three-dimensional solution structure of beta cryptogein, a beta elicitin secreted by a phytopathogenic fungus Phytophthora cryptogea.
Fefeu, S., Bouaziz, S., Huet, J.C., Pernollet, J.C., Guittet, E.(1997) Protein Sci 6: 2279-2284
- PubMed: 9385630
- DOI: https://doi.org/10.1002/pro.5560061101
- Primary Citation of Related Structures:
1BEG - PubMed Abstract:
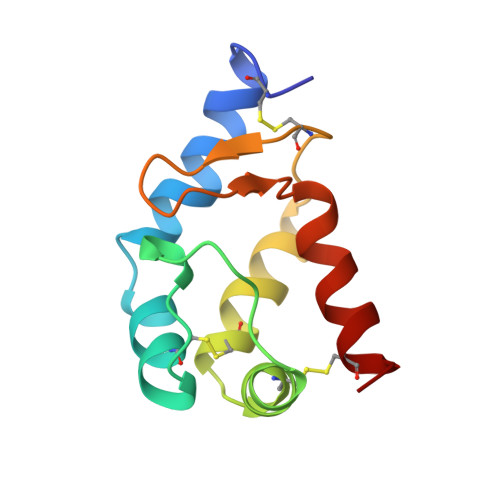
Cryptogein belongs to a new family of 10-kDa proteins called elicitins. Elicitins are necrotic and signaling proteins secreted by Phytophthora spp. responsible for the incompatible reaction and systemic hypersensitive-like necroses of diverse plant species leading to resistance against fungal or bacterial plant pathogens. The solution structure of beta cryptogein from Phytophthora cryptogea fungus was determined by using multidimensional heteronuclear nuclear magnetic resonance spectroscopy. A set of 18 structures was calculated using 1360 NOE-derived distance restraints and 40 dihedral angle restraints obtained from 3JHNH alpha couplings. The RMS deviation from the mean structure is 0.87 +/- 0.14 A for backbone atoms and 1.34 +/- 0.14 A for all the non-hydrogen atoms of residues 2 to 98. The structure of beta cryptogein reveals a novel protein fold, with five helices and a double-stranded beta-sheet facing an omega-loop. One edge of the beta-sheet and the adjacent face of the omega-loop form a hydrophobic cavity. This cavity made of highly conserved residues represents a plausible binding site. Residue 13, which has been identified from directed mutagenesis and natural sequence comparison studies as a key amino acid involved in the differential control of necrosis, is surface exposed and could contribute to the binding to a ligand or a receptor. The solution structure is close to the X-ray structure, with slight differences lightly due to the crystal packing.
Organizational Affiliation:
Laboratoire de RMN, ICSN-CNRS, Gif-sur-Yvette, France.