Structure of Ptr ToxA: An RGD-Containing Host-Selective Toxin from Pyrenophora tritici-repentis
Sarma, G.N., Manning, V.A., Ciuffetti, L.M., Karplus, P.A.(2005) Plant Cell 17: 3190-3202
- PubMed: 16214901
- DOI: https://doi.org/10.1105/tpc.105.034918
- Primary Citation of Related Structures:
1ZLD, 1ZLE - PubMed Abstract:
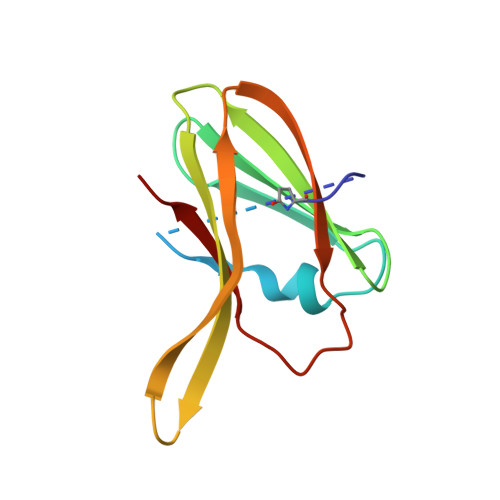
Tan spot of wheat (Triticum aestivum), caused by the fungus Pyrenophora tritici-repentis, has significant agricultural and economic impact. Ptr ToxA (ToxA), the first discovered proteinaceous host-selective toxin, is produced by certain P. tritici-repentis races and is necessary and sufficient to cause cell death in sensitive wheat cultivars. We present here the high-resolution crystal structure of ToxA in two different crystal forms, providing four independent views of the protein. ToxA adopts a single-domain, beta-sandwich fold of novel topology. Mapping of the existing mutation data onto the structure supports the hypothesized importance of an Arg-Gly-Asp (RGD) and surrounding sequence. Its occurrence in a single, solvent-exposed loop in the protein suggests that it is directly involved in recognition events required for ToxA action. Furthermore, the ToxA structure reveals a surprising similarity with the classic mammalian RGD-containing domain, the fibronectin type III (FnIII) domain: the two topologies are related by circular permutation. The similar topologies and the positional conservation of the RGD-containing loop raises the possibility that ToxA is distantly related to mammalian FnIII proteins and that to gain entry it binds to an integrin-like receptor in the plant host.
Organizational Affiliation:
Department of Biochemistry and Biophysics, Oregon State University, Corvallis, Oregon 97331, USA.