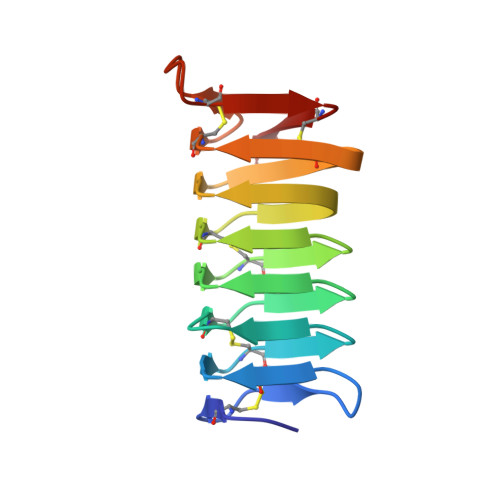
Solution Structure of an Antifreeze Protein CfAFP-501 from Choristoneura fumiferana
Li, C., Guo, X., Jia, Z., Xia, B., Jin, C.(2005) J Biomol NMR 32: 251-256
- PubMed: 16132825
- DOI: https://doi.org/10.1007/s10858-005-8206-3
- Primary Citation of Related Structures:
1Z2F - PubMed Abstract:
Antifreeze proteins (AFPs) are widely employed by various organisms as part of their overwintering survival strategy. AFPs have the unique ability to suppress the freezing point of aqueous solution and inhibit ice recrystallization through binding to the ice seed crystals and restricting their growth. The solution structure of CfAFP-501 from spruce budworm has been determined by NMR spectroscopy. Our result demonstrates that CfAFP-501 retains its rigid and highly regular structure in solution. Overall, the solution structure is similar to the crystal structure except the N- and C-terminal regions. NMR spin-relaxation experiments further indicate the overall rigidity of the protein and identify a collection of residues with greater flexibilities. Furthermore, Pro91 shows a cis conformation in solution instead of the trans conformation determined in the crystal structure.
Organizational Affiliation:
Beijing Nuclear Magnetic Resonance Center, Beijing, China.