A structural basis for allosteric control of DNA recombination by lambda integrase.
Biswas, T., Aihara, H., Radman-Livaja, M., Filman, D., Landy, A., Ellenberger, T.(2005) Nature 435: 1059-1066
- PubMed: 15973401
- DOI: https://doi.org/10.1038/nature03657
- Primary Citation of Related Structures:
1Z19, 1Z1B, 1Z1G - PubMed Abstract:
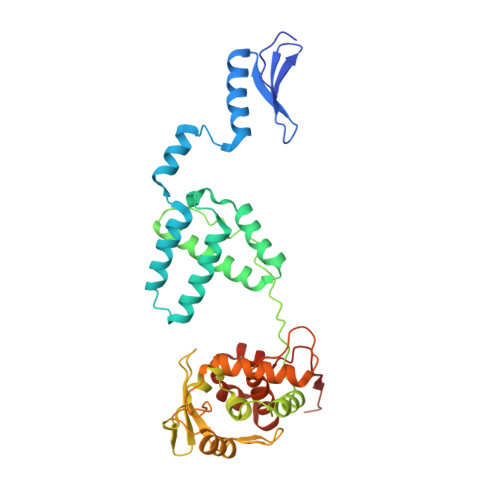
Site-specific DNA recombination is important for basic cellular functions including viral integration, control of gene expression, production of genetic diversity and segregation of newly replicated chromosomes, and is used by bacteriophage lambda to integrate or excise its genome into and out of the host chromosome. lambda recombination is carried out by the bacteriophage-encoded integrase protein (lambda-int) together with accessory DNA sites and associated bending proteins that allow regulation in response to cell physiology. Here we report the crystal structures of lambda-int in higher-order complexes with substrates and regulatory DNAs representing different intermediates along the reaction pathway. The structures show how the simultaneous binding of two separate domains of lambda-int to DNA facilitates synapsis and can specify the order of DNA strand cleavage and exchange. An intertwined layer of amino-terminal domains bound to accessory (arm) DNAs shapes the recombination complex in a way that suggests how arm binding shifts the reaction equilibrium in favour of recombinant products.
Organizational Affiliation:
Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, Massachusetts 02115, USA.