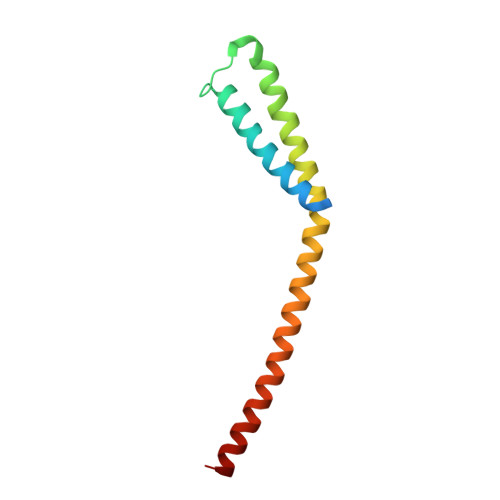
A conserved mediator hinge revealed in the structure of the MED7.MED21 (Med7.Srb7) heterodimer.
Baumli, S., Hoeppner, S., Cramer, P.(2005) J Biol Chem 280: 18171-18178
- PubMed: 15710619
- DOI: https://doi.org/10.1074/jbc.M413466200
- Primary Citation of Related Structures:
1YKE, 1YKH - PubMed Abstract:
The Mediator of transcriptional regulation is the central coactivator that enables a response of RNA polymerase II (Pol II) to activators and repressors. We present the 3.0-A crystal structure of a highly conserved part of the Mediator, the MED7.MED21 (Med7.Srb7) heterodimer. The structure is very extended, spanning one-third of the Mediator length and almost the diameter of Pol II. It shows a four-helix bundle domain and a coiled-coil protrusion connected by a flexible hinge. Four putative protein binding sites on the surface allow for assembly of the Mediator middle module and for binding of the conserved subunit MED6, which is shown to bridge to the Mediator head module. A flexible MED6 bridge and the MED7.MED21 hinge could account for changes in overall Mediator structure upon binding to Pol II or activators. Our results support the idea that transcription regulation involves conformational changes within the general machinery.
Organizational Affiliation:
Gene Center, University of Munich (Ludwig-Maximilians-Universität), Department of Chemistry and Biochemistry, Feodor-Lynen-Strasse 25, 81377 Munich, Germany.