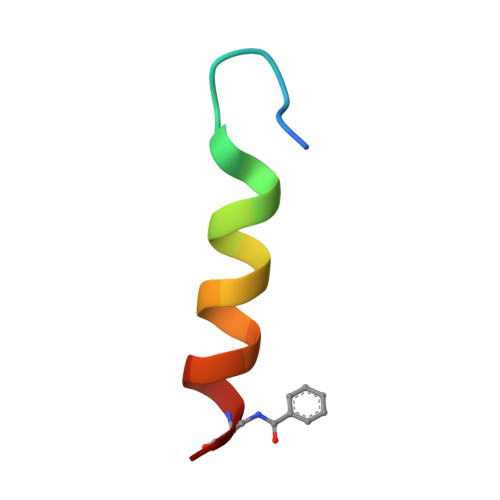
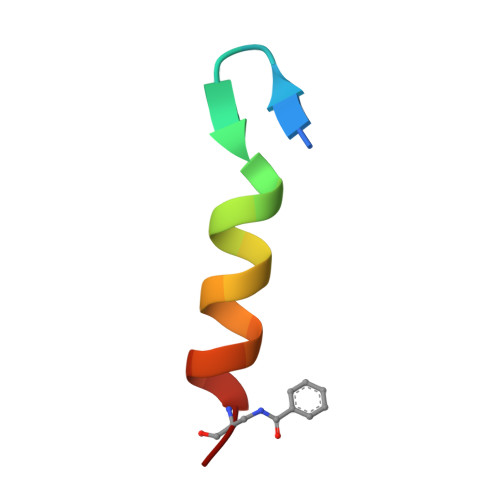
Design of a Heterospecific, Tetrameric, 21-Residue Miniprotein with Mixed alpha/beta Structure.
Ali, M.H., Taylor, C.M., Grigoryan, G., Allen, K.N., Imperiali, B., Keating, A.E.(2005) Structure 13: 225-234
- PubMed: 15698566
- DOI: https://doi.org/10.1016/j.str.2004.12.009
- Primary Citation of Related Structures:
1XOF - PubMed Abstract:
The study of short, autonomously folding peptides, or "miniproteins," is important for advancing our understanding of protein stability and folding specificity. Although many examples of synthetic alpha-helical structures are known, relatively few mixed alpha/beta structures have been successfully designed. Only one mixed-secondary structure oligomer, an alpha/beta homotetramer, has been reported thus far. In this report, we use structural analysis and computational design to convert this homotetramer into the smallest known alpha/beta-heterotetramer. Computational screening of many possible sequence/structure combinations led efficiently to the design of short, 21-residue peptides that fold cooperatively and autonomously into a specific complex in solution. A 1.95 A crystal structure reveals how steric complementarity and charge patterning encode heterospecificity. The first- and second-generation heterotetrameric miniproteins described here will be useful as simple models for the analysis of protein-protein interaction specificity and as structural platforms for the further elaboration of folding and function.
Organizational Affiliation:
Department of Chemistry, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, Massachusetts 02139, USA.