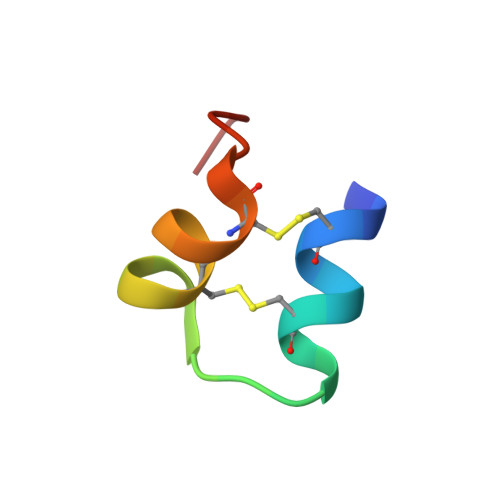
An unusual fold for potassium channel blockers: NMR structure of three toxins from the scorpion Opisthacanthus madagascariensis
Chagot, B., Pimentel, C., Dai, L., Pil, J., Tytgat, J., Nakajima, T., Corzo, G., Darbon, H., Ferrat, G.(2005) Biochem J 388: 263-271
- PubMed: 15631621
- DOI: https://doi.org/10.1042/BJ20041705
- Primary Citation of Related Structures:
1WQC, 1WQD, 1WQE - PubMed Abstract:
The Om-toxins are short peptides (23-27 amino acids) purified from the venom of the scorpion Opisthacanthus madagascariensis. Their pharmacological targets are thought to be potassium channels. Like Csalpha/beta (cystine-stabilized alpha/beta) toxins, the Om-toxins alter the electrophysiological properties of these channels; however, they do not share any sequence similarity with other scorpion toxins. We herein demonstrate by electrophysiological experiments that Om-toxins decrease the amplitude of the K+ current of the rat channels Kv1.1 and Kv1.2, as well as human Kv1.3. We also determine the solution structure of three of the toxins by use of two-dimensional proton NMR techniques followed by distance geometry and molecular dynamics. The structures of these three peptides display an uncommon fold for ion-channel blockers, Csalpha/alpha (cystine-stabilized alpha-helix-loop-helix), i.e. two alpha-helices connected by a loop and stabilized by two disulphide bridges. We compare the structures obtained and the dipole moments resulting from the electrostatic anisotropy of these peptides with those of the only other toxin known to share the same fold, namely kappa-hefutoxin1.
Organizational Affiliation:
Architecture et Fonction des Macromolécules Biologiques, UMR 6098, CNRS et Universités d'Aix-Marseille I et II, 31 Chemin Joseph Aiguier, 13402 Marseille Cedex 20, France.