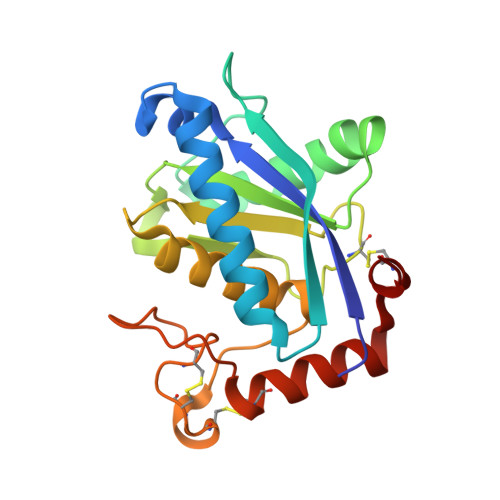
Crystal structure of H2-proteinase from the venom of Trimeresurus flavoviridis.
Kumasaka, T., Yamamoto, M., Moriyama, H., Tanaka, N., Sato, M., Katsube, Y., Yamakawa, Y., Omori-Satoh, T., Iwanaga, S., Ueki, T.(1996) J Biochem 119: 49-57
- PubMed: 8907175
- DOI: https://doi.org/10.1093/oxfordjournals.jbchem.a021215
- Primary Citation of Related Structures:
1WNI - PubMed Abstract:
The crystal structure of the zinc-protease, H2-proteinase, isolated from the venom of Trimeresurus flavoviridis has been determined. The crystallographic R factor is 0.176 for 10,635 reflections with Fobs > 2sigma(Fobs) in the 8.0 to 2.2 Angstrom resolution range. The enzyme has two domains with a cleft in which a catalytic zinc atom is located. The N-terminal domain is composed of four helices around a central five-stranded beta-sheet. The irregularly folded C-terminal domain contains one helix and two disulfide bridges. These two domains are linked by a disulfide bridge. In the zinc environment, the catalytic zinc atom forms a distorted tetrahedral coordination with three histidines and one catalytic water molecule, and the methionine-containing turn is structurally conserved. These are distinctive features of the metzincins, one of the zinc metalloprotease superfamilies. The entire structure shows good agreement with that of two Crotalus snake venom proteases, adamalysin II and atrolysin C. The H2-proteinase, however, contains no structural calcium ions, and differences of disulfide configurations and the coordination of the catalytic water molecule exist as compared with the other two proteases.
Organizational Affiliation:
Department of Life Science, Faculty of Bioscience and Biotechnology, Tokyo Institute of Technology, Midori-ku, Yokohama.