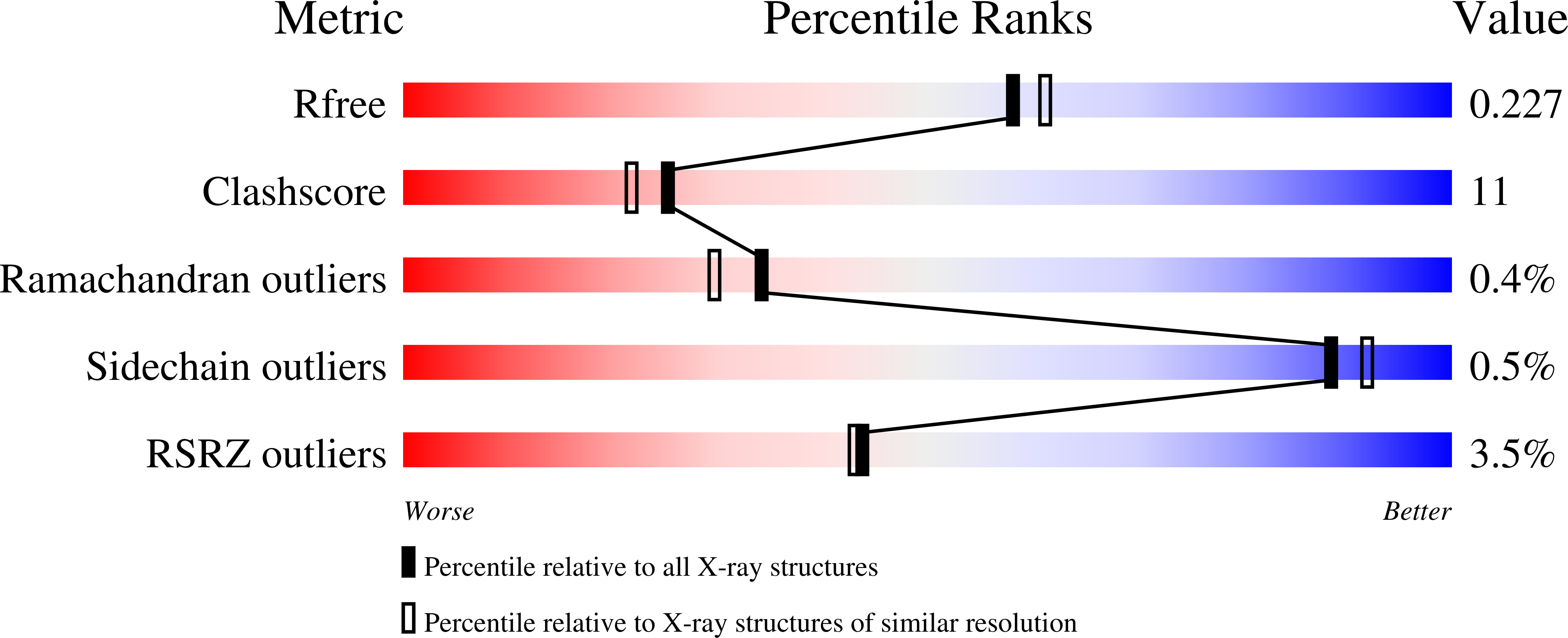
An unexpected phosphate binding site in Glyceraldehyde 3-Phosphate Dehydrogenase: Crystal structures of apo, holo and ternary complex of Cryptosporidium parvum enzyme
Cook, W.J., Senkovich, O., Chattopadhyay, D.(2009) BMC Struct Biol 9: 9-9
- PubMed: 19243605
- DOI: https://doi.org/10.1186/1472-6807-9-9
- Primary Citation of Related Structures:
1VSU, 1VSV, 3CIF - PubMed Abstract:
The structure, function and reaction mechanism of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) have been extensively studied. Based on these studies, three anion binding sites have been identified, one 'Ps' site (for binding the C-3 phosphate of the substrate) and two sites, 'Pi' and 'new Pi', for inorganic phosphate. According to the original flip-flop model, the substrate phosphate group switches from the 'Pi' to the 'Ps' site during the multistep reaction. In light of the discovery of the 'new Pi' site, a modified flip-flop mechanism, in which the C-3 phosphate of the substrate binds to the 'new Pi' site and flips to the 'Ps' site before the hydride transfer, was proposed. An alternative model based on a number of structures of B. stearothermophilus GAPDH ternary complexes (non-covalent and thioacyl intermediate) proposes that in the ternary Michaelis complex the C-3 phosphate binds to the 'Ps' site and flips from the 'Ps' to the 'new Pi' site during or after the redox step.
Organizational Affiliation:
Department of Pathology, University of Alabama at Birmingham, Birmingham, AL 35294, USA. wjcook@uab.edu