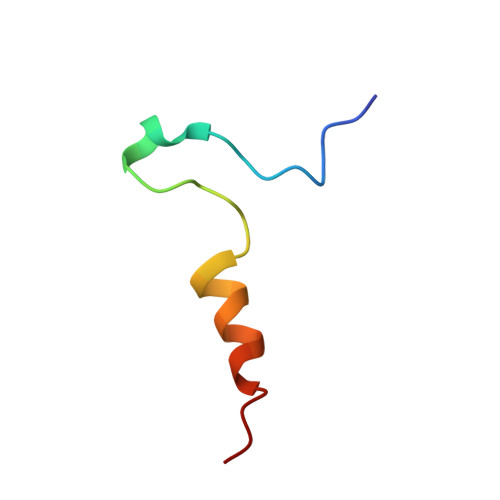
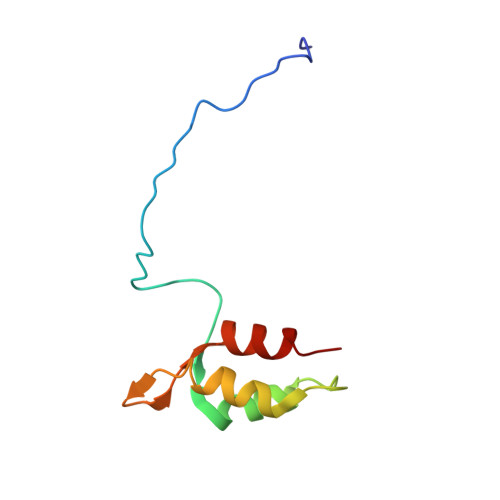
Structural basis for redox regulation of Yap1 transcription factor localization.
Wood, M.J., Storz, G., Tjandra, N.(2004) Nature 430: 917-921
- PubMed: 15318225
- DOI: https://doi.org/10.1038/nature02790
- Primary Citation of Related Structures:
1SSE - PubMed Abstract:
The ability of organisms to alter their gene expression patterns in response to environmental changes is essential for viability. A central regulator of the response to oxidative stress in Saccharomyces cerevisiae is the Yap1 transcription factor. Upon activation by increased levels of reactive oxygen species, Yap1 rapidly redistributes to the nucleus where it regulates the expression of up to 70 genes. Here we identify a redox-regulated domain of Yap1 and determine its high-resolution solution structure. In the active oxidized form, a nuclear export signal (NES) in the carboxy-terminal cysteine-rich domain is masked by disulphide-bond-mediated interactions with a conserved amino-terminal alpha-helix. Point mutations that weaken the hydrophobic interactions between the N-terminal alpha-helix and the C-terminal NES-containing domain abolished redox-regulated changes in subcellular localization of Yap1. Upon reduction of the disulphide bonds, Yap1 undergoes a change to an unstructured conformation that exposes the NES and allows redistribution to the cytoplasm. These results reveal the structural basis of redox-dependent Yap1 localization and provide a previously unknown mechanism of transcription factor regulation by reversible intramolecular disulphide bond formation.
Organizational Affiliation:
Cell Biology and Metabolism Branch, National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, Maryland 20892-5430, USA.