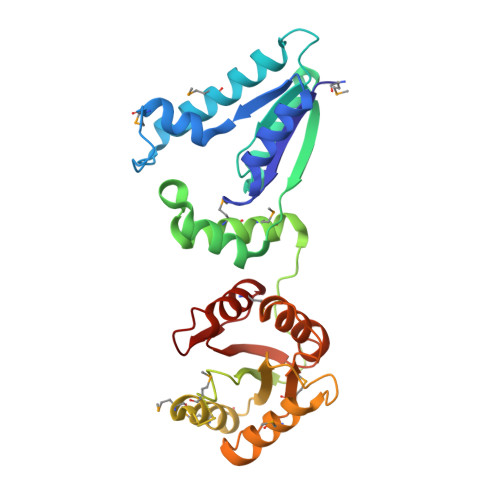
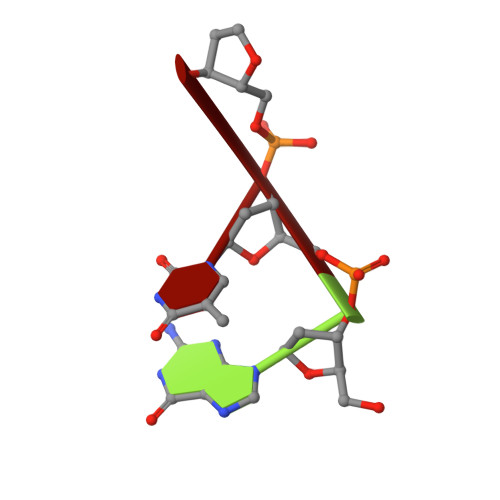
Recognition of DNA substrates by T4 bacteriophage polynucleotide kinase.
Eastberg, J.H., Pelletier, J., Stoddard, B.L.(2004) Nucleic Acids Res 32: 653-660
- PubMed: 14754987
- DOI: https://doi.org/10.1093/nar/gkh212
- Primary Citation of Related Structures:
1RC8, 1RPZ, 1RRC - PubMed Abstract:
T4 phage polynucleotide kinase (PNK) displays 5'-hydroxyl kinase, 3'-phosphatase and 2',3'-cyclic phosphodiesterase activities. The enzyme phosphorylates the 5' hydroxyl termini of a wide variety of nucleic acid substrates, a behavior studied here through the determination of a series of crystal structures with single-stranded (ss)DNA oligonucleotide substrates of various lengths and sequences. In these structures, the 5' ribose hydroxyl is buried in the kinase active site in proper alignment for phosphoryl transfer. Depending on the ssDNA length, the first two or three nucleotide bases are well ordered. Numerous contacts are made both to the phosphoribosyl backbone and to the ordered bases. The position, side chain contacts and internucleotide stacking interactions of the ordered bases are strikingly different for a 5'-GT DNA end than for a 5'-TG end. The base preferences displayed at those positions by PNK are attributable to differences in the enzyme binding interactions and in the DNA conformation for each unique substrate molecule.
Organizational Affiliation:
Fred Hutchinson Cancer Research Center and the Graduate Program in Molecular and Cellular Biology, University of Washington, Seattle, WA 98109, USA.