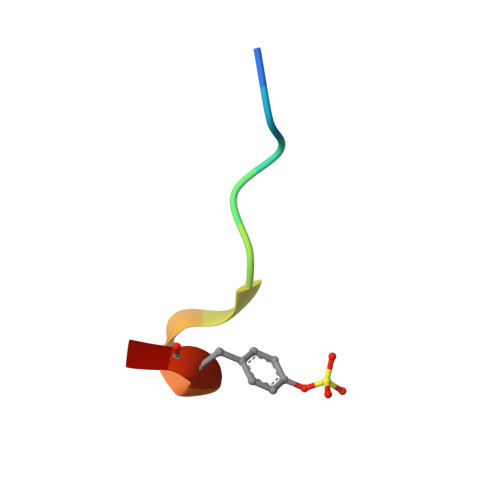
The N-acyloxyiminium ion aza-Prins route to octahydroindoles: total synthesis and structural confirmation of the antithrombotic marine natural product oscillarin
Hanessian, S., Tremblay, M., Petersen, J.F.W.(2004) J Am Chem Soc 126: 6064-6071
- PubMed: 15137772
- DOI: https://doi.org/10.1021/ja030669g
- Primary Citation of Related Structures:
1RIW - PubMed Abstract:
The first enantiocontrolled total synthesis of the marine natural product oscillarin is described. The proposed structure and absolute configuration of oscillarin is thus confirmed, and a previously assigned structure of a subunit was shown to be incorrect. The X-ray structure of an oscillarin-thrombin complex was resolved at 2.0 A resolution, which validated its potent inhibitory activity against the enzyme with an IC(50) = 28 nM. Methodology was developed for the synthesis of enantiopure octahydroindole-2-carboxylic acids with usable functionality at C-6. The method consists of the halocarbocyclization of N-acyloxyiminium ions containing an olefinic tether in the presence of tin tetrachloride or tin tetrabromide. This N-acyloxyiminium ion aza-Prins carbocyclization proved to be general for the construction of octahydroindole and perhydroquinoline 2-carboxylic acids. Mechanistic rationales are based on an antiperiplanar attack of the terminal alkene on the iminium ion, leading to an incipient secondary carbocation which is trapped by halide via an equatorial attack. X-ray crystal structures of products corroborate the expected stereochemistry.
Organizational Affiliation:
Department of Chemistry, Université de Montréal, C.P. 6128, Station Centre-ville, Montréal, P.Q., H3C 3J7 Canada. stephen.hanessian@umontreal.ca