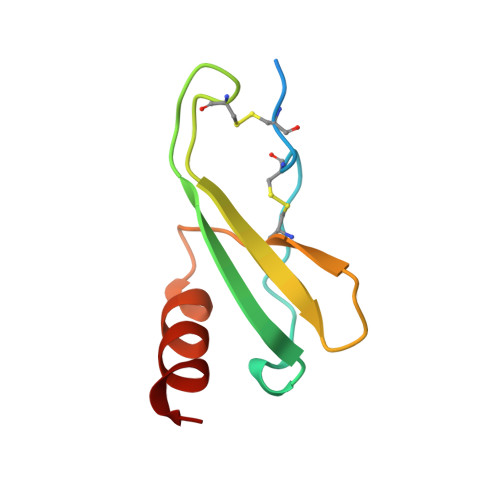
The three-dimensional structure of bovine platelet factor 4 at 3.0-A resolution.
St Charles, R., Walz, D.A., Edwards, B.F.(1989) J Biol Chem 264: 2092-2099
- PubMed: 2914894
- Primary Citation of Related Structures:
1PLF - PubMed Abstract:
Platelet factor 4 (PF4), which is released by platelets during coagulation, binds very tightly to negatively charged oligosaccharides such as heparin. To date, six other proteins are known that are homologous in sequence with PF4 but have quite different functions. The structure of a tetramer of bovine PF4 complexed with one Ni(CN)4(2-) molecule has been determined at 3.0 A resolution and refined to an R factor of 0.28. The current model contains residues 24-85, no solvent, and one overall temperature factor. Residues 1-13, which carried an oligosaccharide chain, were removed with elastase to induce crystallization; residues 14-23 and presumably 86-88 are disordered in the electron density map. Because no heavy atom derivative was isomorphous with the native crystals, the complex of PF4 with one Ni(CN)4(2-) molecule was solved using a single, highly isomorphous Pt(CN)4(2-) derivative and the iterative, single isomorphous replacement method. The secondary structure of the PF4 subunit, from amino- to carboxyl-terminal end, consists of an extended loop, three strands of antiparallel beta-sheet arranged in a Greek key, and one alpha-helix. The tetramer contains two extended, six-stranded beta-sheets, each formed by two subunits, which are arranged back-to-back to form a "beta-bilayer" structure with two buried salt bridges sandwiched in the middle. The carboxyl-terminal alpha-helices, which contain lysine residues that are thought to be intimately involved in binding heparin, are arranged as antiparallel pairs on the surface of each extended beta-sheet.
Organizational Affiliation:
Department of Biochemistry, Wayne State University School of Medicine, Detroit, Michigan 48201.