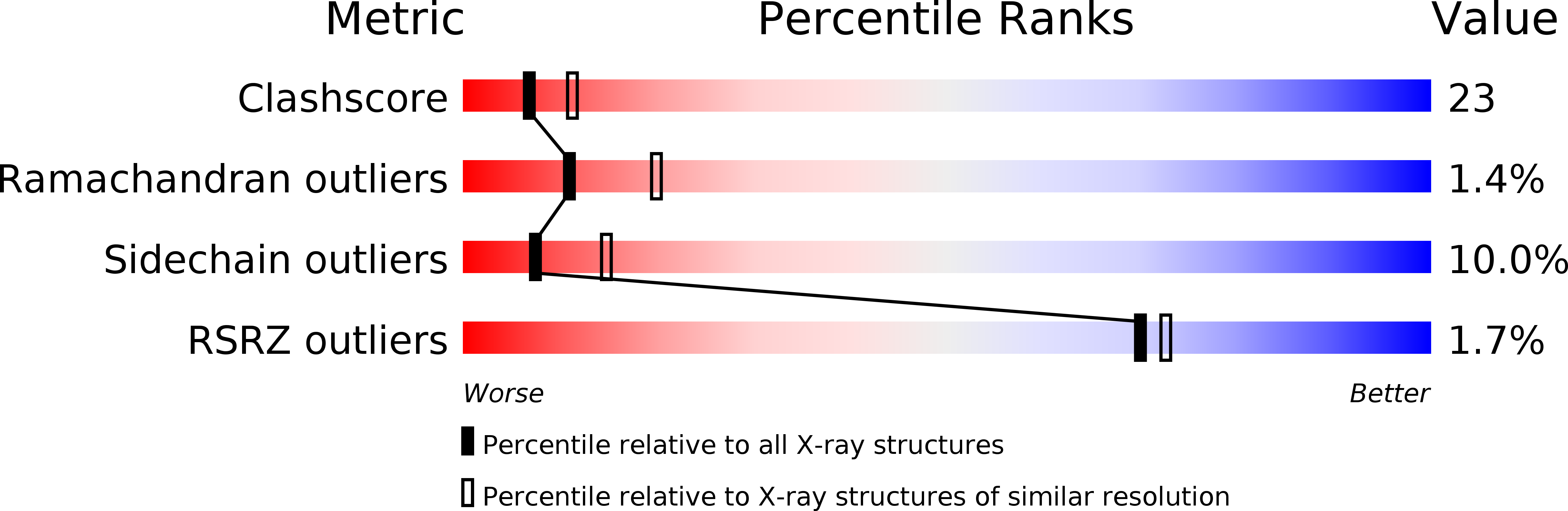Haemoglobin of the antarctic fish Pagothenia bernacchii. Amino acid sequence, oxygen equilibria and crystal structure of its carbonmonoxy derivative.
Camardella, L., Caruso, C., D'Avino, R., di Prisco, G., Rutigliano, B., Tamburrini, M., Fermi, G., Perutz, M.F.(1992) J Mol Biol 224: 449-460
- PubMed: 1560461
- DOI: https://doi.org/10.1016/0022-2836(92)91007-c
- Primary Citation of Related Structures:
1PBX - PubMed Abstract:
The Antarctic fish Pagothenia bernacchii has one major haemoglobin, Hb1 (over 95% of the total blood content). Hb1 has a strong alkaline Bohr effect and at low pH exhibits the reduced ligand affinity and co-operativity that comprise the Root effect. We have determined the complete amino acid sequence of P. bernacchii Hb1 and also the structure of its carbonmonoxy derivative by X-ray crystallography, to a resolution of 2.5 A. The crystallographic R-factor of the refined structure is 18%. The three-dimensional structure of this fish haemoglobin is similar to that of human haemoglobin A, with a root-mean-square difference in main-chain atom positions of 1.4 A after superimposition of the two structures, despite only 48% homology of their amino acid sequences (including insertion of a single residue in the CD region of the fish alpha-chain). Large structural differences occur only at the N and C termini of both the alpha- and beta-chains. Neither these nor other smaller structural differences provide any obvious explanation of the Root effect of this or other fish haemoglobins.
Organizational Affiliation:
Institute of Protein Biochemistry and Enzymology, C.N.R., Naples, Italy.