Crystal Structure of APC1665, YUAD protein from Bacillus subtilis
Kim, Y., Joachimiak, A., Edwards, A., Skarina, T., Savchenko, A.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| yuaD protein | 195 | Bacillus subtilis | Mutation(s): 6 | 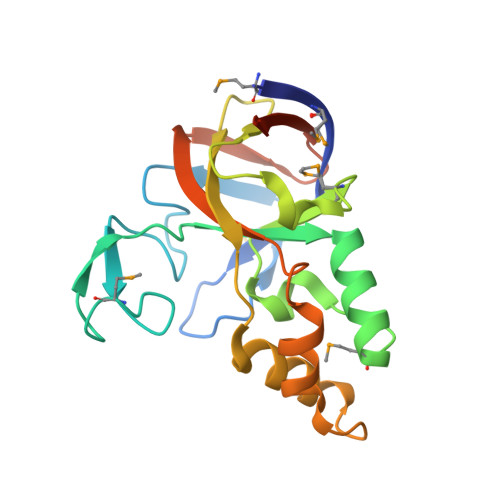 | |
UniProt | |||||
Find proteins for O32079 (Bacillus subtilis (strain 168)) Explore O32079 Go to UniProtKB: O32079 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O32079 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SO4 Query on SO4 | C [auth A], F [auth B], G [auth B] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| CL Query on CL | D [auth A], E [auth A], H [auth B] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MSE Query on MSE | A, B | L-PEPTIDE LINKING | C5 H11 N O2 Se |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 42.652 | α = 90 |
| b = 87.202 | β = 90 |
| c = 122.249 | γ = 90 |
| Software Name | Purpose |
|---|---|
| CNS | refinement |
| d*TREK | data reduction |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| CNS | phasing |