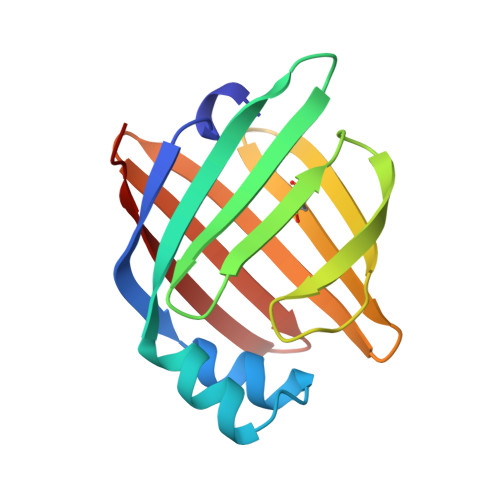
The Crystal Structure of Echinococcus Granulosus Fatty-Acid-Binding Protein 1
Jakobsson, E., Alvite, G., Bergfors, T., Esteves, A., Kleywegt, G.J.(2003) Biochim Biophys Acta 1649: 40
- PubMed: 12818189
- DOI: https://doi.org/10.1016/s1570-9639(03)00151-1
- Primary Citation of Related Structures:
1O8V - PubMed Abstract:
We describe the 1.6 A crystal structure of the fatty-acid-binding protein EgFABP1 from the parasitic platyhelminth Echinococcus granulosus. E. granulosus causes hydatid disease, which is a major zoonosis. EgFABP1 has been implicated in the acquisition, storage, and transport of lipids, and may be important to the organism since it is incapable of synthesising most of its lipids de novo. Moreover, EgFABP1 is a promising candidate for a vaccine against hydatid disease. The crystal structure reveals that EgFABP1 has the expected 10-stranded beta-barrel fold typical of the family of intracellular lipid-binding proteins, and that it is structurally most similar to P2 myelin protein. We describe the comparison of the crystal structure of EgFABP1 with these proteins and with an older homology model for EgFABP1. The electron density reveals the presence of a bound ligand inside the cavity, which we have interpreted as palmitic acid. The carboxylate group of the fatty acid interacts with the protein's P2 motif, consisting of a conserved triad R em leader R-x-Y. The hydrophobic tail of the ligand assumes a fairly flat, U-shaped conformation and has relatively few interactions with the protein.We discuss some of the structural implications of the crystal structure of EgFABP1 for related platyhelminthic FABPs.
Organizational Affiliation:
Department of Cell and Molecular Biology, Uppsala University, Biomedical Centre, Box 596, SE-751 24, Uppsala, Sweden.