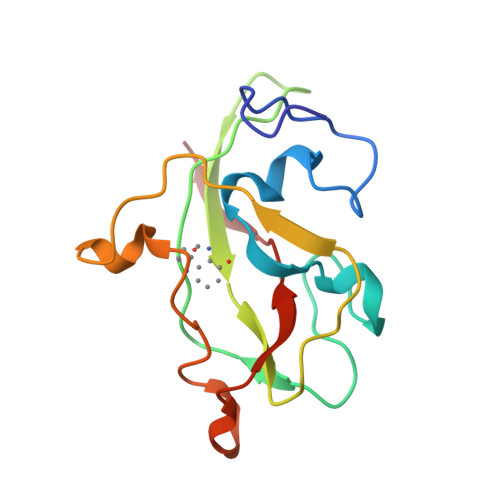
Active site structure of methylamine dehydrogenase: hydrazines identify C6 as the reactive site of the tryptophan-derived quinone cofactor.
Huizinga, E.G., van Zanten, B.A., Duine, J.A., Jongejan, J.A., Huitema, F., Wilson, K.S., Hol, W.G.(1992) Biochemistry 31: 9789-9795
- PubMed: 1390754
- DOI: https://doi.org/10.1021/bi00155a036
- Primary Citation of Related Structures:
1MAE, 1MAF, 2MAD - PubMed Abstract:
To identify the reactive part of the orthoquinone function of the tryptophan-derived cofactor found in methylamine dehydrogenase (MADH), we have determined the crystal structures of MADH from Thiobacillus versutus inhibited by methylhydrazine and (2,2,2-trifluoroethyl)hydrazine. Extra electron density attached to C6 of the tryptophyl tryptophanquinone cofactor shows that this atom and not C7 is the reactive part of the ortho-quinone moiety. The density retained after hydrazine inhibition is much less extensive than expected, however, suggesting that partial breakdown of the inhibitors after reaction with the cofactor may take place. A detailed description is presented of the cofactor environment in an improved model of MADH which now includes information from the recently determined gene sequence of the cofactor-containing subunit [Ubbink, M., van Kleef, M.A.G., Kleinjan, D., Hoitink, C.W.G., Huitema, F., Beintema, J.J., Duine, J.A., & Canters, G.W. (1991) Eur. J. Biochem. 202, 1003-1012]. We hypothesize that Asp76 is responsible for proton abstraction from the alpha-carbon of the substrate during catalysis.
Organizational Affiliation:
Department of Chemistry, Groningen University, The Netherlands.