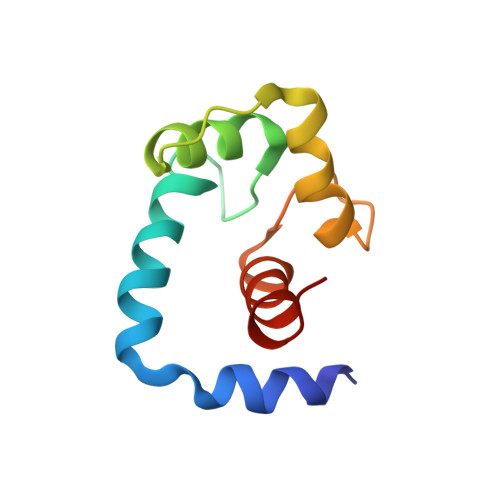
Structure of the regulatory N-domain of human cardiac troponin C in complex with human cardiac troponin I147-163 and bepridil.
Wang, X., Li, M.X., Sykes, B.D.(2002) J Biol Chem 277: 31124-31133
- PubMed: 12060657
- DOI: https://doi.org/10.1074/jbc.M203896200
- Primary Citation of Related Structures:
1LXF - PubMed Abstract:
Cardiac troponin C (cTnC) is the Ca(2+)-dependent switch for contraction in heart muscle and a potential target for drugs in the therapy of heart failure. Ca(2+) binding to the regulatory domain of cTnC (cNTnC) induces little structural change but sets the stage for cTnI binding. A large "closed" to "open" conformational transition occurs in the regulatory domain upon binding cTnI(147-163) or bepridil. This raises the question of whether cTnI(147-163) and bepridil compete for cNTnC.Ca(2+). In this work, we used two-dimensional (1)H,(15)N-heteronuclear single quantum coherence (HSQC) NMR spectroscopy to examine the binding of bepridil to cNTnC.Ca(2+) in the absence and presence of cTnI(147-163) and of cTnI(147-163) to cNTnC.Ca(2+) in the absence and presence of bepridil. The results show that bepridil and cTnI(147-163) bind cNTnC.Ca(2+) simultaneously but with negative cooperativity. The affinity of cTnI(147-163) for cNTnC.Ca(2+) is reduced approximately 3.5-fold by bepridil and vice versa. Using multinuclear and multidimensional NMR spectroscopy, we have determined the structure of the cNTnC.Ca(2+).cTnI(147-163).bepridil ternary complex. The structure reveals a binding site for cTnI(147-163) primarily located on the A/B interhelical interface and a binding site for bepridil in the hydrophobic pocket of cNTnC.Ca(2+). In the structure, the N terminus of the peptide clashes with part of the bepridil molecule, which explains the negative cooperativity between cTnI(147-163) and bepridil for cNTnC.Ca(2+). This structure provides insights into the features that are important for the design of cTnC-specific cardiotonic drugs, which may be used to modulate the Ca(2+) sensitivity of the myofilaments in heart muscle contraction.
Organizational Affiliation:
Canadian Institutes for Health Research Group in Protein Structure and Function, Department of Biochemistry, University of Alberta, Edmonton, Alberta T6G 2H7, Canada.