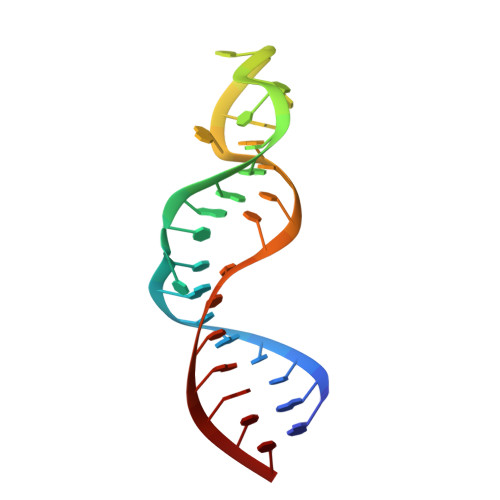
Structure of TAR RNA complexed with a Tat-TAR interaction nanomolar inhibitor that was identified by computational screening
Du, Z., Lind, K.E., James, T.L.(2002) Chem Biol 9: 707-712
- PubMed: 12079782
- DOI: https://doi.org/10.1016/s1074-5521(02)00151-5
- Primary Citation of Related Structures:
1LVJ - PubMed Abstract:
HIV-1 TAR RNA functions critically in viral replication by binding the transactivating regulatory protein Tat. We recently identified several compounds that experimentally inhibit the Tat-TAR interaction completely at a 100 nM concentration. We used computational screening of the 181,000-compound Available Chemicals Directory against the three-dimensional structure of TAR [1]. Here we report the NMR-derived structure of TAR complexed with acetylpromazine. This structure represents a new class of compounds with good bioavailability and low toxicity that bind with high affinity to TAR. NMR data unambiguously show that acetylpromazine binds only to the unique 5' bulge site to which the Tat protein binds. Specificity and affinity of binding are conferred primarily by a network of base stacking and hydrophobic interactions. Acetylpromazine alters the structure of free TAR less than Tat peptides and neomycin do.
Organizational Affiliation:
Department of Pharmaceutical Chemistry, University of California, San Francisco, San Franciso, CA 94143, USA.