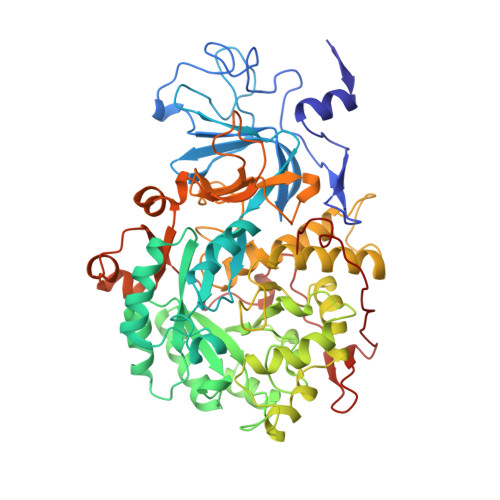
Structures of the Klebsiella aerogenes urease apoenzyme and two active-site mutants.
Jabri, E., Karplus, P.A.(1996) Biochemistry 35: 10616-10626
- PubMed: 8718850
- DOI: https://doi.org/10.1021/bi960424z
- Primary Citation of Related Structures:
1KRA, 1KRB, 1KRC - PubMed Abstract:
Urease from Klebsiella aerogenes [Jabri et al. (1995) Science 268, 998-1004] is an (alpha beta gamma)3 trimer with each alpha-subunit having an (alpha beta)8-barrel domain containing a binickel active center. Here we examine structure-function relations for urease in more detail through structural analysis of the urease apoenzyme at 2.3 A resolution and mutants of two key catalytic residues (H219A and H320A) at 2.5 A resolution. With the exception of the active site, in which a water molecule takes the place of the missing carbamate and nickel atoms, the structure of the apoenzyme is nearly identical to that of the holoenzyme, suggesting a high degree of preorganization which helps explain the tight binding of nickel. In the structure of H219A, the major change involves a conformational shift and ordering of the active site flap, but a small shift in the side chain of Asp alpha 221 could contribute to the lower activity of H219A. In the H320A structure, the catalytic water, primarily a Ni-2 ligand in the holoenzyme, shifts into a bridging position. This shift shows that the nickel ligation is rather sensitive to the environment and the change in ligation may contribute to the 10(5)-fold lower activity of H320A. In addition, these results show that urease is resilient to the loss of nickel ions and mutations. Analysis of the urease tertiary/quaternary structure suggests that the stability of this enzyme may be largely due to its burial of an unusually large fraction of its residues: 50% in the gamma-subunit, 30% in the beta-subunit, and 60% in the alpha-subunit.
Organizational Affiliation:
Section of Biochemistry, Molecular and Cell Biology, Cornell University, Ithaca, New York 14853, USA.