Homonuclear (1)H-NMR assignment and structural characterization of human immunodeficiency virus type 1 Tat Mal protein.
Gregoire, C., Peloponese Jr., J.M., Esquieu, D., Opi, S., Campbell, G., Solomiac, M., Lebrun, E., Lebreton, J., Loret, E.P.(2001) Biopolymers 62: 324-335
- PubMed: 11857271
- DOI: https://doi.org/10.1002/bip.10000
- Primary Citation of Related Structures:
1K5K - PubMed Abstract:
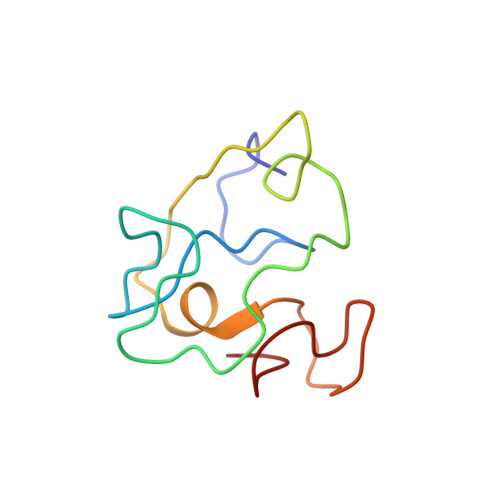
The transacting transcriptional activator (Tat) is a viral protein essential for activation of the human immunodeficiency virus (HIV) genes, and it plays an important role in HIV induced immunodeficiency. We report the NMR structural characterization of the active Tat Mal variant that belongs to a highly virulent D-subtype HIV type-1 (HIV-1) strain (Mal) found mainly in Africa. A full Tat Mal protein (87 residues) is synthesized. This synthetic protein is active in a transactivation assay with HeLa cells infected with the HIV long terminal repeated noncoding sequences of the HIV-1 provirus (LTR) lac Z gene. Homonuclear (1)H-NMR spectra allows the sequential assignment of the Tat Mal spin systems. Simulating annealing generates 20 conformers with similar folding. The geometry of the mean structure is optimized with energy minimization to obtain a final structure. As the European variant (Tat Bru) the N-terminal region of Tat Mal constitutes the core, and there is a hydrophobic pocket composed of the conserved Trp 11 interacting with several aromatic residues. The two functional regions of Tat (basic and the cysteine-rich regions) are well exposed to the solvent. A short alpha-helix is observed in region V adjacent to the basic region. This alpha helix induces local structural variations compared to the NMR structure of Tat Bru, and it brings the cysteine-rich and basic regions closer. This study suggests that similar folding exists among Tat variants.
Organizational Affiliation:
Laboratoire d'Ingénierie des Systèmes Macromoléculaires, Institut de Biologie Structurale et Microbiologie, CNRS UPR 9027, 31 Chemin Joseph Aiguier, 13402 Marseille, France.