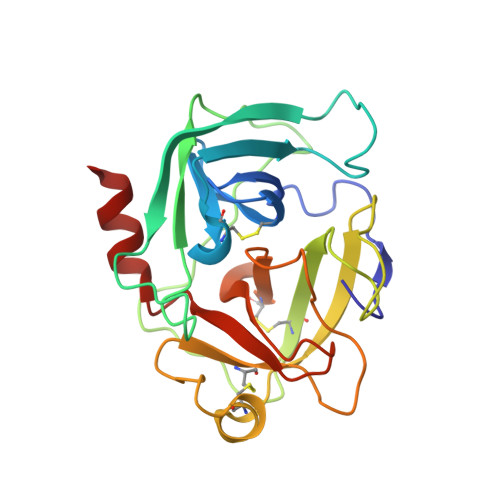
The three-dimensional structure of human granzyme B compared to caspase-3, key mediators of cell death with cleavage specificity for aspartic acid in P1.
Rotonda, J., Garcia-Calvo, M., Bull, H.G., Geissler, W.M., McKeever, B.M., Willoughby, C.A., Thornberry, N.A., Becker, J.W.(2001) Chem Biol 8: 357-368
- PubMed: 11325591
- DOI: https://doi.org/10.1016/s1074-5521(01)00018-7
- Primary Citation of Related Structures:
1IAU - PubMed Abstract:
Granzyme B, one of the most abundant granzymes in cytotoxic T-lymphocyte (CTL) granules, and members of the caspase (cysteine aspartyl proteinases) family have a unique cleavage specificity for aspartic acid in P1 and play critical roles in the biochemical events that culminate in cell death.
Organizational Affiliation:
Department of Endocrinology and Chemical Biology, Merck Research Laboratories, Rahway, NJ 07065-0900, USA. rotonda@merck.com