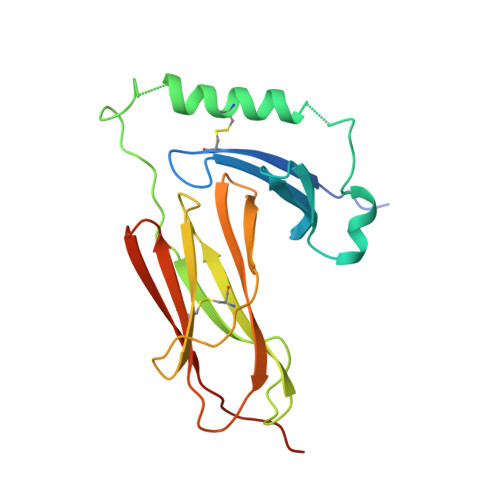
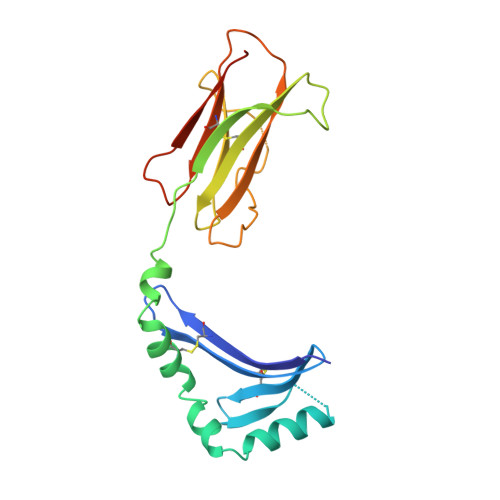
The structure of HLA-DM, the peptide exchange catalyst that loads antigen onto class II MHC molecules during antigen presentation.
Mosyak, L., Zaller, D.M., Wiley, D.C.(1998) Immunity 9: 377-383
- PubMed: 9768757
- DOI: https://doi.org/10.1016/s1074-7613(00)80620-2
- Primary Citation of Related Structures:
1HDM - PubMed Abstract:
The three-dimensional structure of the soluble ecto-domain of HLA-DM has been determined to 2.5 A resolution by X-ray crystallography. HLA-DM has both peptide exchange activity and acts as a chaperone to peptide-free class II MHC molecules. As predicted, the structure is similar to that of classical class II MHC molecules except that the peptide-binding site is altered to an almost fully closed groove. An unusual cavity is found at the center of the region that binds peptides in class II MHC molecules, and a tryptophanrich lateral surface is identified that is a candidate both for binding to HLA-DR, to effect catalysis, and to HLA-DO, an inhibitor.
Organizational Affiliation:
Department of Cellular and Molecular Biology, Howard Hughes Medical Institute, Harvard University, Cambridge, Massachusetts 02138, USA. mosyak@crystal.harvard.edu