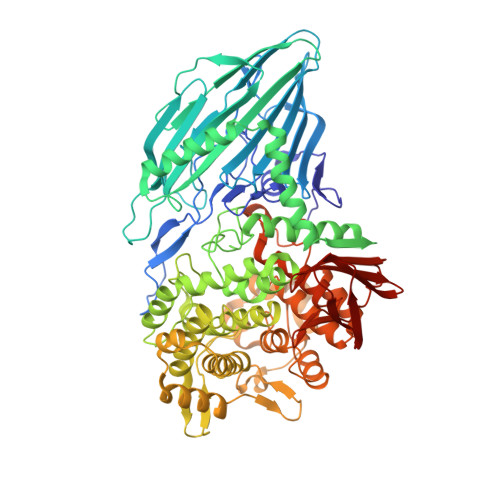
Crystal Structure of Maltose Phosphorylase from Lactobacillus Brevis: Unexpected Evolutionary Relationship with Glucoamylases.
Egloff, M.-P., Uppenberg, J., Haalck, L., Van Tilbeurgh, H.(2001) Structure 9: 689
- PubMed: 11587643
- DOI: https://doi.org/10.1016/s0969-2126(01)00626-8
- PubMed Abstract:
Maltose phosphorylase (MP) is a dimeric enzyme that catalyzes the conversion of maltose and inorganic phosphate into beta-D-glucose-1-phosphate and glucose without requiring any cofactors, such as pyridoxal phosphate. The enzyme is part of operons that are involved in maltose/malto-oligosaccharide metabolism. Maltose phosphorylases have been classified in family 65 of the glycoside hydrolases. No structure is available for any member of this family.
Organizational Affiliation:
Architecture et Fonction des Macromolécules Biologiques, Centre National de la Recherche Scientifique, Unite Mixte de Recherche 6098, Université d'Aix-Marseille, I et II, Case 925, 13288, Marseille, France.